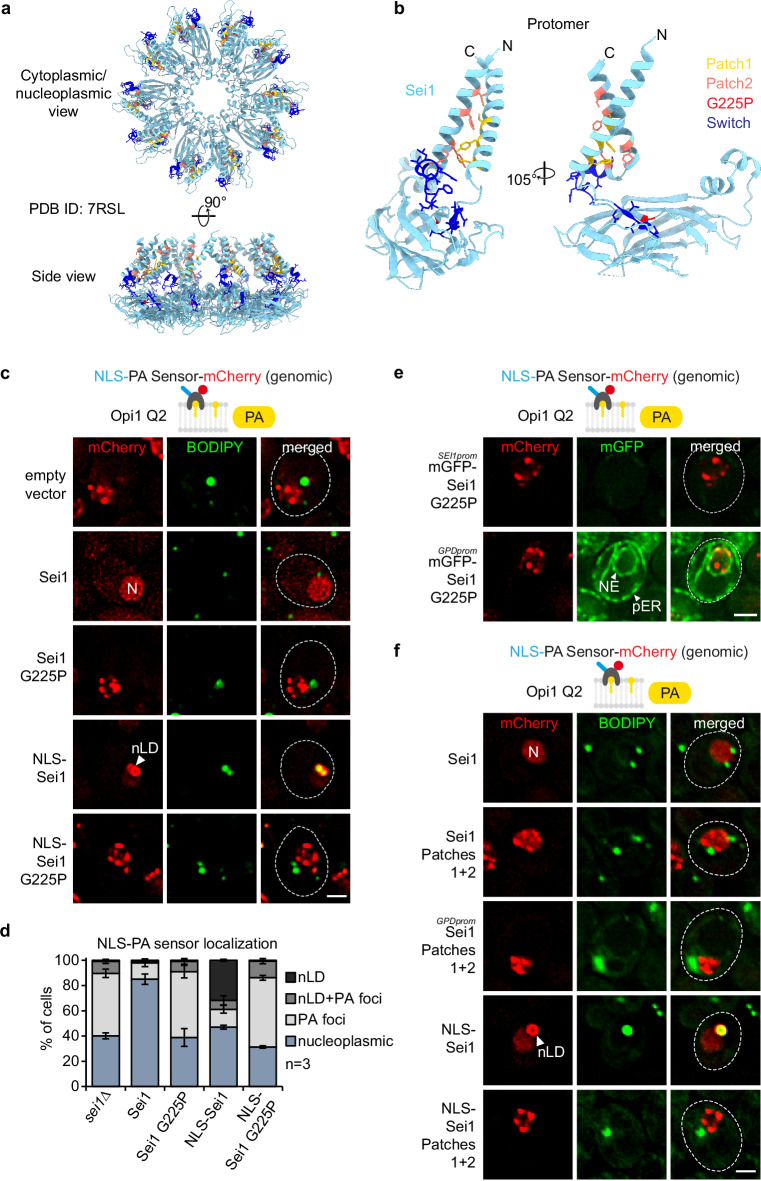
Fig. 5. PA defects at the INM in Sei1 lipodystrophy and TM contact mutants.
a Cartoon representation of S. cerevisiae Sei1 homodecamer (PDB ID: 7RSL) with select amino acid residues shown in atom (stick) representation. The amino acid residues, which were either substituted or deleted in corresponding Sei1 mutants, are colour-coded (yellow: Patch1, salmon: Patch2, red: G225P, blue: Switch). Note that the mutated amino acid residues do not map to the inter-subunit interfaces, and are therefore unlikely to disrupt the formation of the Sei1 ring assembly. b Close-up view of the Sei1 protomer (PDB ID: 7RSL, B conformation) shown in hybrid cartoon-atom representation as in Fig. 5a. c Live imaging of sei1∆ cells expressing genomically integrated NLS-PA-mCherry sensor and the indicated plasmid-based mGFP-SEI1 constructs. BODIPY stains LDs. nLDs have a BODIPY-positive core surrounded by a PA-rich shell. Note that even though cells contain mGFP-Sei1, the green BODIPY fluorescence signal is significantly brighter, hence Sei1 fluorescence remains undetectable when the settings for BODIPY imaging are applied. N, nucleus; nLD, nuclear lipid droplet. Scale bar, 2 μm. d Quantification of NLS-PA-mCherry sensor localization in (c). Mean value and standard deviation indicated. n, number of biological replicates. More than 435 cells analysed for each condition. Source data are provided as a Source Data file. e Live imaging of sei1∆ cells expressing genomically integrated NLS-PA-mCherry sensor and plasmid-based mGFP-sei1 G225P constructs from the endogenous SEI1 or a strong GPD promoter. Scale bar, 2 μm. f Live imaging of sei1∆ cells expressing genomically integrated NLS-PA-mCherry sensor and indicated plasmid-based mGFP-SEI1 constructs from the endogenous SEI1 or a strong GPD promoter. BODIPY stains LDs. N, nucleus; nLD, nuclear lipid droplet. Scale bar, 2 μm.