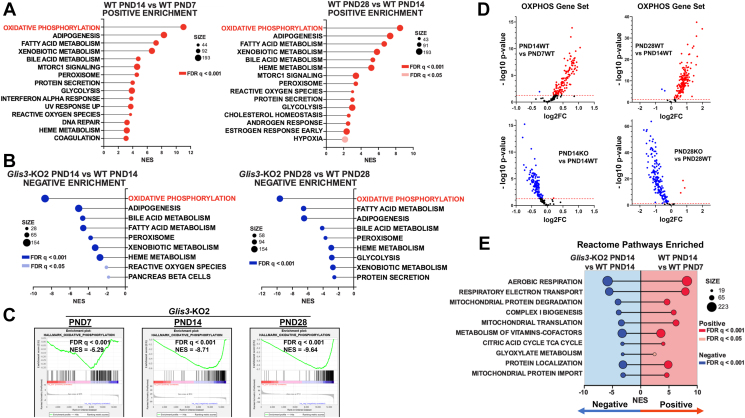
Figure 1.
Mitochondrial-related pathways are suppressed in Glis3-KO2 kidneys. (A) GSEA Hallmark pathway showing the top positively enriched age-associated pathways for WT kidneys ranked by the normalized enrichment score (NES) and showing FDR and gene set size. The topmost enriched pathways are in red. (B) GSEA Hallmark pathway showing the top negatively enriched age-associated pathways for Glis3-KO2 kidneys ranked by NES and showing FDR and gene set size. The topmost enriched pathways are in blue. (C) GSEA OXPHOS enrichment plots showing negative enrichment in the Glis3-KO2 kidneys at PND7, 14, and 28. (D) Volcano plots of all Hallmark OXPHOS genes showing upregulation with age (red) in the WT PND14 vs WT PND7 and WT PND28 vs WT PND14 and downregulation of the OXPHOS gene set (blue) in Glis3-KO2 PND14 vs WT PND14 and Glis3-KO2 vs WT PND28 kidneys. (E) GSEA pathway analysis depicting the top enriched Reactome pathways in the WT PND14 vs WT PND7 compared to the Glis3-KO2 PND14 vs WT PND14 showing opposite enrichment. For WT the pathways show positive enrichment, and for the Glis3-KO2 showing negative enrichment. Reactome pathways ranked by NES in the Glis3-KO2 PND14, and showing FDR and gene set size. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)