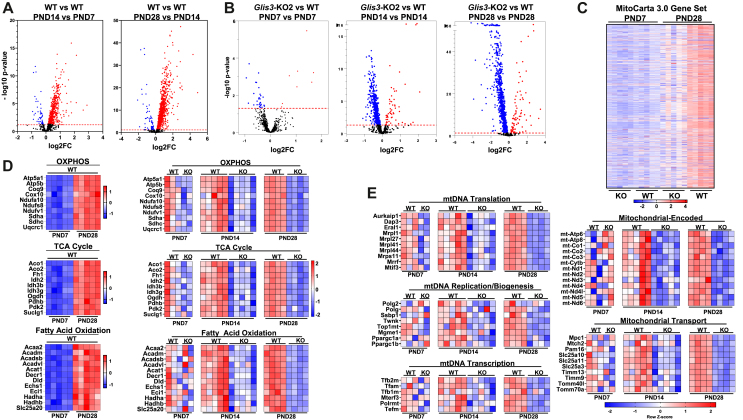
Figure 2.
The expression of mitochondria-related genes is decreased in Glis3-KO2 kidneys. (A) Visualization of differential gene expression in WT kidneys. Volcano plots of DEGS of WT PND14 vs WT PND7 and WT PND28 vs WT PND14 kidney RNA-seq data against the MitoCarta 3.0 gene dataset. (B) Visualization of differential gene expression in Glis3-KO2 kidneys. Volcano plots of DEGS of PND7, 14, and 28 Glis3-KO2 and WT kidney RNA-seq data against the MitoCarta 3.0 gene dataset. (C) Heatmap of the MitoCarta 3.0 gene set in alphabetical order. Gene expression is compared between PND7 and PND28 WT and Glis3-KO2. Upregulated genes are represented in red and downregulated genes in blue. Expression values are shown as z-scores of the rlog-transformed values for each gene. (D, E) Heatmaps of DEGs in Glis3-KO2 versus WT kidneys at PND7, 14, and 28 labeled with mitochondria-associated pathways. For OXPHOS, TCA Cycle, and FAO a heat map of WT PND28 vs WT PND7 is included showing the increase in expression with age. Upregulated genes are represented in red and downregulated genes in blue. Expression values are shown as z-scores of the rlog-transformed values for each gene. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)