Figure 1.
High-throughput drug combination screens with ATRi and TROP2 expression in patient biopsies and cell lines
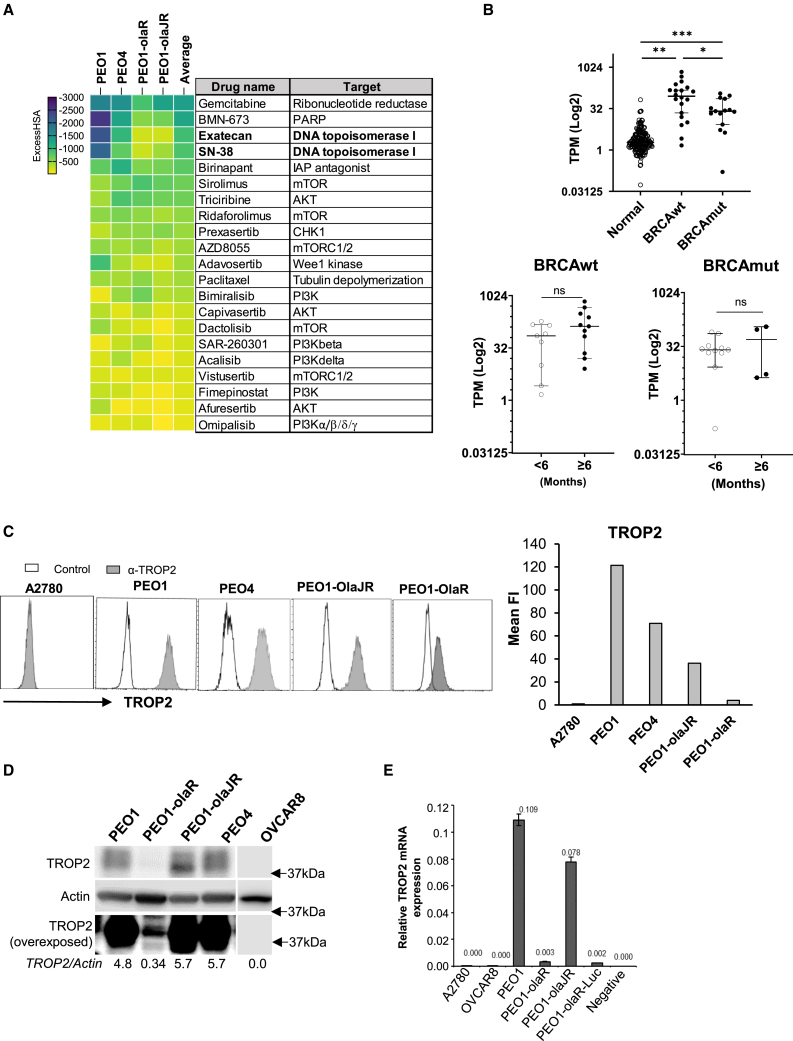
(A) The heatmap is a ranked average ExcessHSA from a 10 × 10 combination screen of ATRi (ceralasertib) with various drugs selected from a 6 × 6 screen (Figure S1), in PARPi-sensitive PEO1, PARPi-resistant PEO1-olaJR, and PEO1-olaR and de novo PARPi-resistant PEO4. This experiment was performed once.
(B) Expression values (Log2 transcripts per million, TPM) for TROP2 mRNA (TACSTD2) from both BRCA-mutant (BRCAmut) cohort (n = 15) and BRCA wild-type (BRCAwt) cohort (n = 20) of patients were compared against TPM values of TROP2 from normal ovarian tissues (normal) obtained from the GTEx database (n = 180). TROP2 values for the no-clinical benefit (<6 months PFS) or clinical benefit (≥6 months PFS) groups for cohorts BRCAwt (n = 9 and n = 11 respectively) and BRCAmut (n = 11 and n = 4 respectively) were compared as well. Graphs show TROP2 expression as median TPM (Log2) ± 95% confidence interval (CI) for each cohort or group. Statistical significance was determined using the unpaired Mann-Whitney U-test and is shown as p-values ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, ns, not significant.
(C) Flow cytometric analysis for TROP2 expression (gray) was performed after incubating cells with recombinant anti-TROP2 hRS7 antibody followed by goat anti-human F(ab)2-AF488 secondary antibody as given in STAR Methods. In control group (white), cells were treated only with secondary antibody. A2780 cell line was used as a TROP2-negative control. Graphs show the value of mean fluorescence intensity (Mean FI) of TROP2 expression peaks divided by that of control peak (n = >8000 events) for each cell line. Figure is representative of 3 biologically independent experiments.
(D) Immunoblotting was performed to screen the baseline levels of TROP2 protein in the same cell lines. Blots were overexposed (bottom panel) to highlight low TROP2 expression in PEO1-olaR. Densitometric quantitation of TROP2 relative to Actin are included below. Closest molecular weight marker positions are shown on the right. Figure is representative of 3 biologically independent experiments.
(E) Quantitative PCR (qPCR) analysis of total RNA from both TROP2 positive and negative cell lines. Graph is representative of 2 biologically independent experiments and shows TROP2 expression values normalized against that of GAPDH for each cell line. A negative control without RNA template is also included as quality control. Data are represented as mean ± SD.