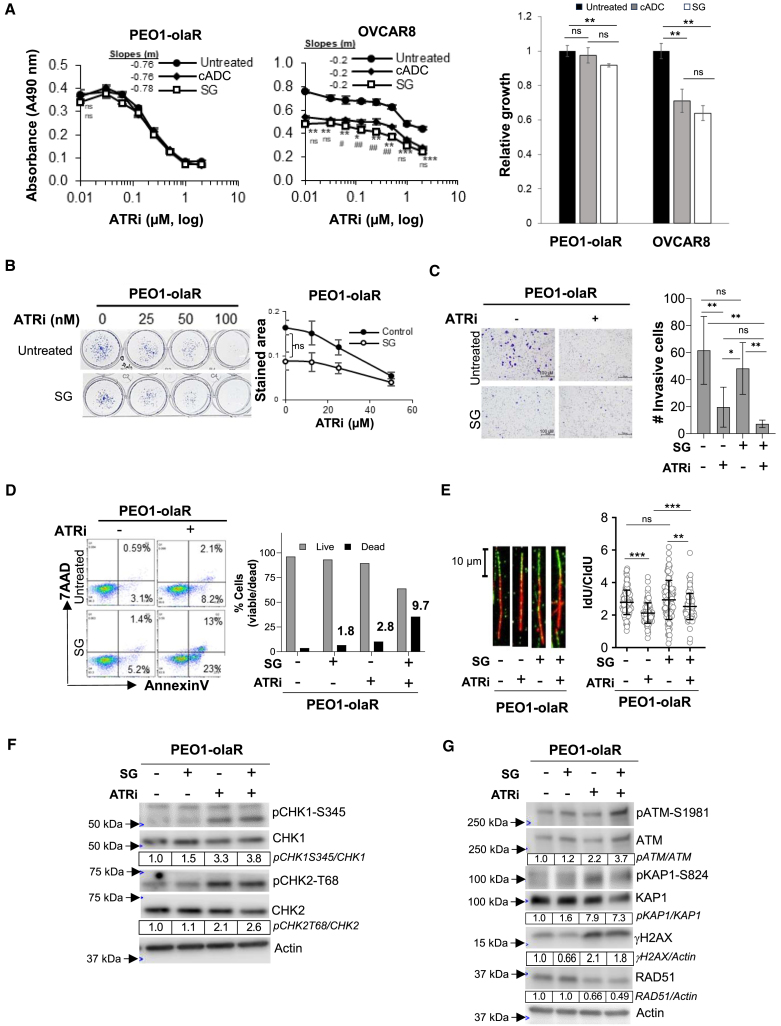
Figure 6.
SG activity in TROP2-negative OVCAR8 and TROP2-low PEO1-olaR
Cells were left untreated or pretreated with SG or cADC (10 μg/mL, 30 min at 37°C and washed thrice with PBS) prior to use in all experiments below. Student’s t test was used to measure significance in all experiments below.
(A) Graphs show mean ± SD (n = 3 biological replicates per treatment) cell growth inhibition from XTT assays for TROP2-negative OVCAR8 and TROP2-low PEO1-olaR cells that were untreated or pretreated with either SG or cADC followed by treatment with a gradient of ATRi (0–10 μM) over 48 h. Untreated cell values (0 μM ATRi) are plotted at 0.01 μM to enable representation on the log X axis. Statistical significance (p-value) was determined by Student’s t test. p-values between untreated and SG plots are indicated by asterisks (∗) while between SG and cADC plots are indicated by hashes (#). Slopes for each curve are derived from linear regression formula “y = mx+b” where y = Y-intercept, x = ATRi (μM) and b = correlation coefficient, calculated from the linear part of the respective curves.
(B) Clonogenic assays were performed over 7 days with untreated or SG pretreated cells against increasing concentrations of berzosertib (ATRi). Representative images are shown on the left while graphs on the right show mean ± SD from 3 biological replicates.
(C) Representative images from one set of Matrigel plates after crystal-violet staining are shown. Following appropriate treatments, invaded cells on underside of inserts were quantified after crystal-violet staining using ImageJ (Fiji) software as detailed in STAR Methods. The plots show mean ± SD for 3 biological replicates. Statistical significance was determined by Student’s t test. Scale bar is 100 μm.
(D) Flow cytometric analysis of viability (AnnexinV/7-AAD) for PEO1-olaR treated with SG or ATRi monotherapy or a combination of both. Dead and live cell proportions are plotted on the right as bar charts from >8000 events. Numbers above bars indicate fold increase in dead cells for each treatment relative to untreated. Results are representative of 3 biologically independent experiments.
(E) DNA fiber assays was performed as explained in STAR Methods. Lengths of both red (IdU) and green (CldU) segments of each strand (∼200 DNA strands per treatment) were measured using Fiji software, and the mean ± SD of the ratio of IdU/CldU from all strands (n = ∼200) were plotted as column charts using GraphPad prism. Representative strands are shown to the left from 2 biologically independent experiments. Statistical significance was determined by Student’s t test. Scale bar is 10 μm.
(F and G) Western blot analysis of lysates from cells pretreated with SG alone or followed by overnight treatment with ATRi berzosertib (1 μM) and analyzed for DNA damage and cell cycle markers. Densitometric quantitation was performed as detailed in the STAR Methods. Densitometric values of phosphorylated proteins relative to total proteins are further normalized to untreated (value of 1) for each cell line and shown below respective images. Values were rounded to two significant numbers. Representative of 3 biologically independent experiments. For panels A, B, C and E, p-values are ∗, #p < 0.05, ∗∗, ##p < 0.01, and ∗∗∗, ###p < 0.001, ns, not significant.