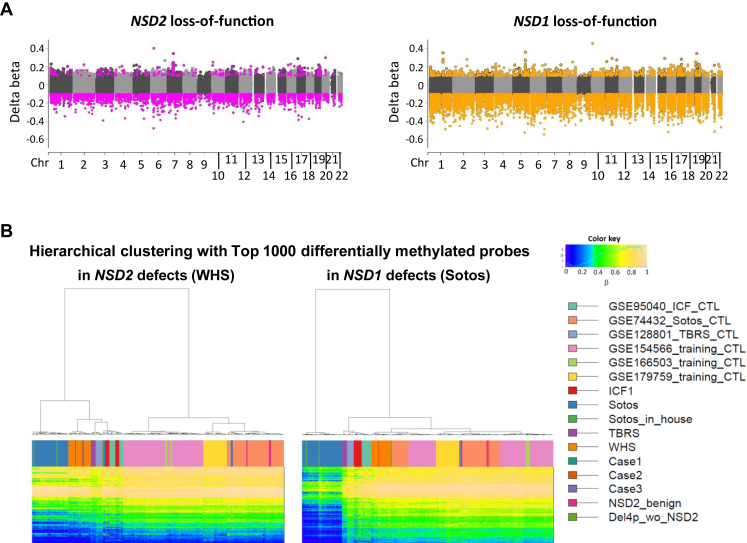
Figure 5.
DNA methylation changes in NSD2 loss-of-function variants and other syndromes. A. Comparison of genome-wide DNA methylation changes between NSD2 and NSD1 loss-of-function variants in peripheral blood cells. Pink and orange dots indicate 3402 and 15,327 probes with significantly differential methylation in NSD2 and NSD1 loss-of-function variants, respectively, with Bonferroni-corrected P < .05 and |delta beta| > 0.15. B. Hierarchical clustering of individuals with NSD2 defects (WHS and Cases), NSD1 defects (Sotos and Sotos inHouse), DNMT3A defects (TBRS), DNMT3B defects (ICF1), and controls of each study by methylation levels of the top 1000 P values DMP probes in NSD2 or in NSD1 defects.