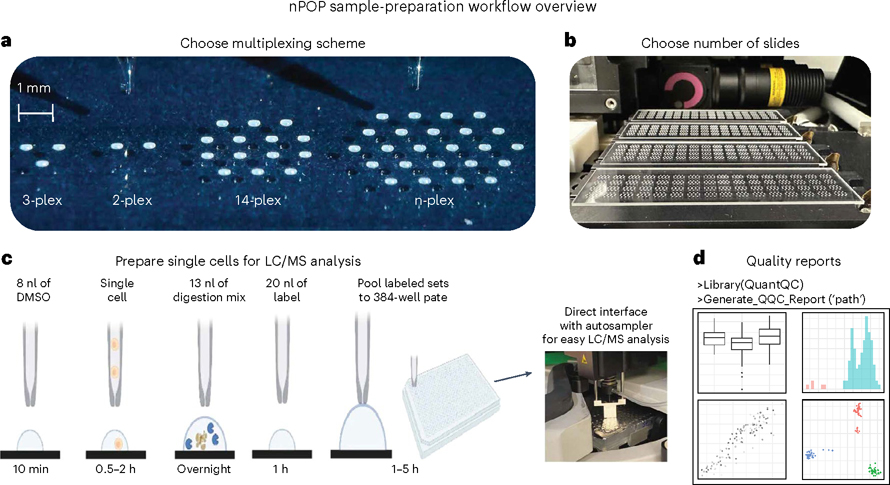
Fig. 1 |. nPOP workflow.
a, nPOP is a proteomic sample-preparation method that prepares single cells in droplets on the surface of fluorocarbon-coated glass slides. This allows for flexible design that can fit any desired multiplexing scheme as reflected by the number of droplets per cluster. b, A picture of a workflow using four glass slides and the 14-plex design allowing simultaneous preparation of 3,584 single cells for prioritized proteomic analysis. c, A schematic of the nPOP method illustrates the steps of cell lysis, protein digestion, peptide labeling, quenching of labeling reaction, sample pooling and transfer of the pooled samples to an autosampler plate. These steps are performed for each single cell (corresponding to a single droplet). d, To analyze data generated from an nPOP sample preparation, the QuantQC R package can be used to map all metadata and generate quality reports for quick evaluation of the experiment. DMSO, dimethyl sulfoxide.