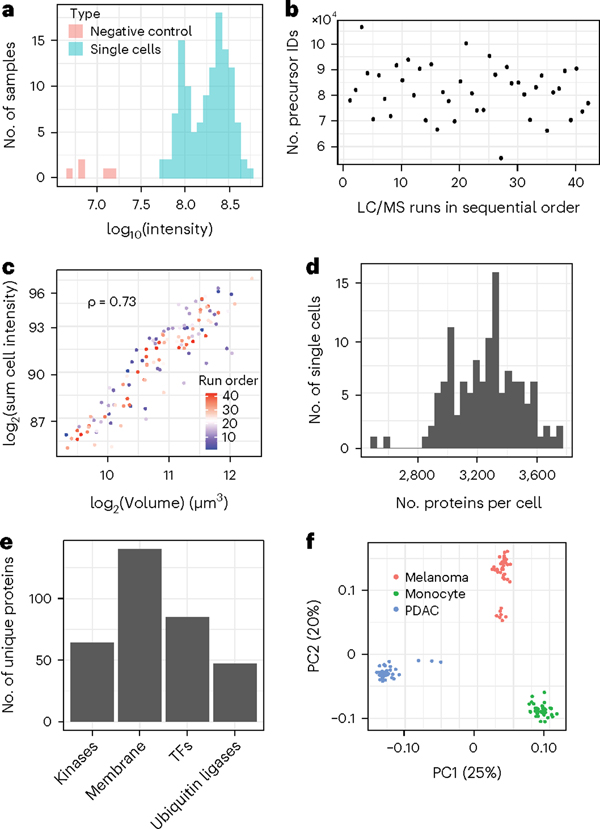
Fig. 5 |. Summary of plexDIA 3-plex data prepared by nPOP.
a, Distribution of the total signal (estimated as summed intensity from all peptides) for both single cells and negative controls, which have received trypsin and label but no cell. b, Number of identified precursors over the course of the LC-MS/MS runs. The numbers remained stable, indicating stable data acquisition. The amount of protein injected depends on the size of the cells in the set; cell size can vary substantially. c, Cell volume has strong positive correlation with summed peptide signal as a proxy for total protein content, indicating consistency of sample preparation. d, Distribution of the number of proteins quantified per cell. e, Number of kinases, membrane proteins, transcription factors (TFs) and ubiquitin ligases identified in the plexDIA data set. f, Principal component analysis shows that cells discretely cluster by cell type. The two clusters of melanoma cells correspond to previously characterized subpopulations in this cell line7.