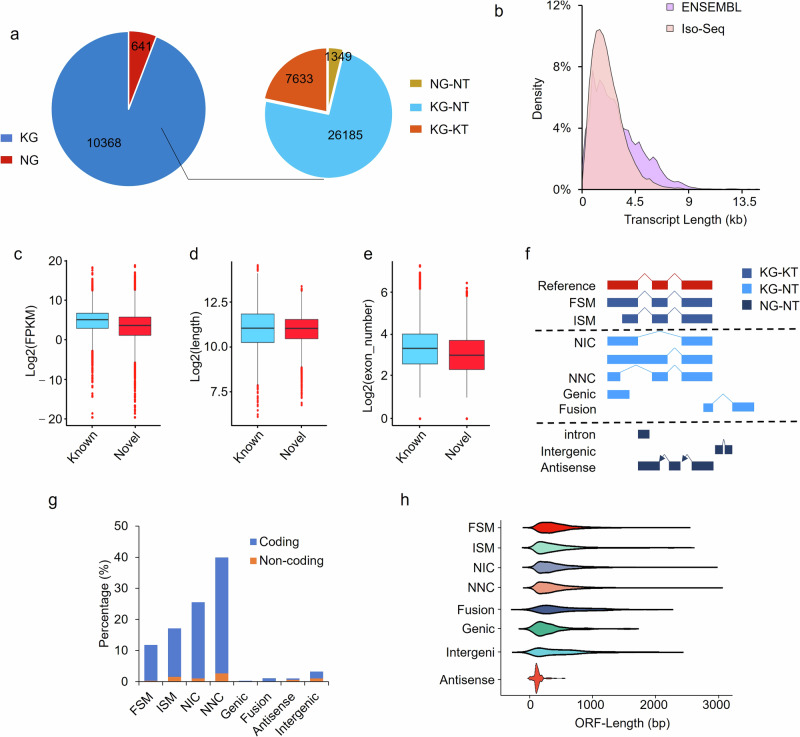
Fig. 1. Characterization of novel transcripts by Iso-Seq.
a Classification of genes identified. NG, novel genes. KG, known genes. NT, novel transcripts. KT, known transcripts. b Length distribution of transcripts. Differences in the abundance (c), length (d) and exon number (e) between novel and known transcripts identified by Cupcake program. f SQANTI3 program classifies the transcripts into nine categories. FSM, full-splice match. ISM, incomplete splice match. NIC, novel in catalog. NNC, novel not in catalog. g In-depth classification of Iso-Seq transcripts by SQANTI3 program. h Length distribution of open reading frames of transcripts among categories.