Abstract
Extracellular vesicles (EVs) are known to mediate cell communications and shape tumor microenvironment. Compared to the well-studied small EVs, the function of large microvesicles (MVs) during tumorigenesis is poorly understood. Here we show the proteome of MVs in Epstein-Barr virus (EBV)-associated gastric cancer (EBVaGC), and identify olfactomedin 4 (OLFM4) is induced by EBV infection and secreted via MVs to promote tumor progression through Hippo signaling. Specifically, OLFM4 is a target gene of the cGAS-STING pathway, and EBV infection activates cGAS-STING pathway and increases OLFM4 expression. Moreover, MV-carried OLFM4 binds with the extracellular cadherin domain of FAT1, thereby impairing its intracellular interaction with MST1 and leading to YAP activation in recipient cells. Together, our study not only reveals a regulatory mechanism though which viral infection is coupled via MVs with intercellular control of the Hippo signaling, but also highlights the OLFM4-Hippo axis as a therapeutic target for EBV-associated cancers.
Subject terms: Cancer microenvironment, Gastric cancer
The functions of microvesicles (MVs) in gastric cancer are less studied compared to other types of extracellular vesicles. Here, the authors report that in Epstein-Barr virus (EBV)-associated gastric cancer (EBVaGC), Olfactomedin 4 protein is enriched in secreted MVs to promote EBVaGC progression by inhibiting the Hippo signaling in tumor cells.
Introduction
Epstein-Barr virus (EBV) is a common virus with an infection rate of over 90% in the adult population1. Infection of EBV has been associated with many types of cancers, such as Hodgkin’s lymphoma, nasopharyngeal carcinoma, diffuse large B-cell lymphoma, and gastric cancer (GC)2. Although EBV can directly invade normal gastric epithelial cells, B lymphocyte-mediated cell-cell contact is thought as a major event for EBV to enter epithelial cells3. EBV-associated gastric cancer (EBVaGC) mostly occurs in the proximal site of the stomach4, and its global incidence rate is ∼10% of gastric carcinomas5. To date, the exact pathological role of EBV in GC is not fully understood.
Extracellular vesicles (EVs), such as exosome (small EVs, 30–150 nm) and microvesicles (MV, large EVs, 50–1000 nm)6,7, are important carriers for cell-cell communications, which play important roles in shaping tissue microenvironment for tumor initiation and progression8–13. Previous studies have shown that exosomes derived from EBV-infected cells carry viral factors such as EBER, LMP1, and host factors such as EGFR and PI3K14,15; and once absorbed by recipient cells, these factors can facilitate carcinogenesis16,17. Compared with exosomes, MVs have larger particle size, and contains more glycosylated and phosphorylated proteins18. Currently, the roles of MVs in tumorigenesis, such as EBVaGC, remains to be clarified.
It is well established that, in response to virus infection, cGAS and its downstream signaling effector STING act as viral DNA sensors in the host cell to activate IRF3- and/or NF-κB-induced interferon expression and thus mediate host defense against viruses19. Recently, the cGAS-STING pathway has been also implicated in cancer immunity and autoimmune diseases20,21. Although it has been speculated that EBV infection may regulate tumorigenesis by activation of the cGAS-STING pathway in the infected cells, it remains unclear how EBV-induced cGAS-STING signaling would affect the tumor microenvironment in an autocrine or paracrine manner. Also, the direct target gene of the cGAS-STING pathway, as well as possible roles in MVs is obscure in the context of EBV-associated cancers.
The evolutionarily conserved Hippo signaling pathway plays important roles in the regulation of organ size, tissue regeneration, immune response and tumorigenesis22,23. The mammalian Hippo pathway can be simplified as an MST1/2-LATS1/2 kinases-mediated phosphorylation cascade that controls subcellular localization of YAP/TAZ, the major downstream transcriptional coactivators that may enter the nucleus and bind TEAD family of transcriptional factors to induce target gene’s expression22,24,25. Compared to the well-studied intracellular signal transduction of the Hippo pathway, the transmembrane and extracellular machinery governing the Hippo pathway remains obscure. In this regard, several transmembrane proteins were identified as cell-surface regulator for Hippo pathway, such as FAT1, GPCRs, and KIRREL126–28. Among them, FAT1 is thought to assemble a multimeric signalome facilitating activation of core Hippo kinases29. And dysregulation of FAT1 has been associated with tumor initiation, progression, invasiveness, stemness and metastasis through the induction of a hybrid EMT state30. However, the extracellular proteins or ligands that relay environmental signals to Fat1 across the plasma membrane remains elusive, even though some membrane proteins such as KK-LC-131, and secreted proteins such as ISLR and Netrin-132,33 have been reported to regulate the Hippo pathway.
Olfactomedin 4 (OLFM4, also known as GW112), a secreted glycoprotein, was first cloned from human hematopoietic myeloid cells34. OLFM4 has been reported to play important roles in cell proliferation, cell adhesion35,36, and has been implicated in many types of cancers especially gastrointestinal cancers37–39. In fact, OLFM4 are widely used as molecular marker for intestinal stem cell and sometimes for cancer stem cell40,41. However, the pathological function of OLFM4 in tumorigenesis including GC development remains to be clarified, especially regarding, (1) upstream signals and mechanisms through which OLFM4 expression and secretion are induced; (2) receptors of OLFM4 and downstream signal transduction across the recipient cell membrane.
Here, we show the proteome of MVs derived from EBVaGC and identify OLFM4 as an MV-carried extracellular inhibitor for the Hippo signaling pathway in recipient cells. Importantly, we reveal that OLFM4 is a direct target gene of the cGAS-STING pathway. Upon EBV infection, the expression and secretion of OLFM4 are significantly increased, and these increased expression of OLFM4 is positively correlated with EBV infection in EBVaGC. When delivered by MVs onto the surface of recipient cells, OLFM4 can interact with FAT1 cadherin domain and act as an extracellular inhibitory ligand to abrogate the intracellular interaction of FAT1 with MST1, thereby activating YAP to promote cell proliferation. Thus, this study describes a regulatory mechanism though which viral infection is coupled via MV-carried OLFM4 with intercellular regulation of the Hippo pathway.
Results
Proteomic profiling of MVs derived from EBVaGC
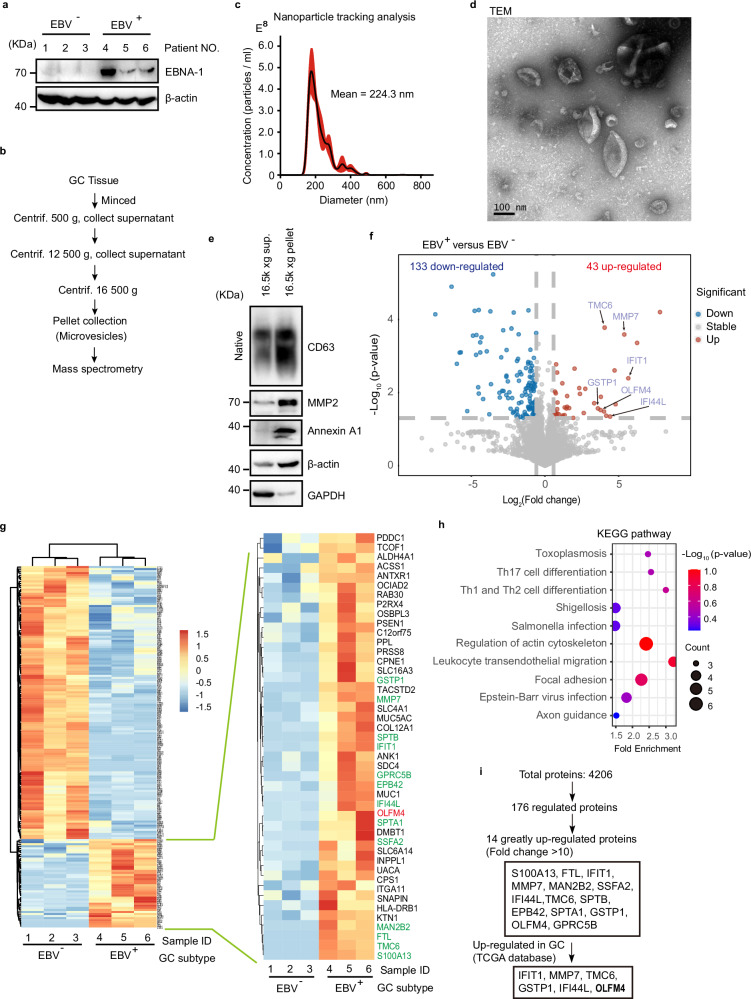
To systematically investigate protein components of MVs in EBVaGC, we collected clinical tissue samples of 3 EBV+ and 3 EBV- gastric cancer patients as indicated by Epstein-Barr virus nuclear antigen 1 (EBNA-1) (Fig. 1a), and isolated MVs from these samples by differential centrifugation (Fig. 1b). Subsequent nanoparticle tracking analysis determined a mean diameter of 224.3 nm for the extracted MVs (Fig. 1c); and transmission electron microscopy (TEM) revealed a spindle-like morphology (Fig. 1d and Supplementary Fig. 1). Further western blot confirmed the specific existence of molecular markers for MVs including Annexin A1 and MMP2 (Fig. 1e).
Fig. 1. Quantitative proteome study of MVs from EBVaGC.
a Western blotting to validate EBV infection of GC tissues used for MV extraction. EBV+ and EBV− human GC tumor samples are used for protein extraction and western blot. EBNA-1, Epstein-Barr virus (EBV)-encoded nuclear antigen 1, a marker protein of EBV. All lanes are loaded with 50 μg of total protein. b Centrifugation protocol and workflow for microvesicle enrichment from gastric cancer tissues. c Representative nanoparticle tracking analysis. d Transmission electron microscopy (TEM) imaging of MVs from gastric cancer tissues. Scale bar represents 100 nm. e Western blot of MVs isolated from gastric cancer for conventional extracellular vesicle markers. GAPDH glyceraldehyde-3-phosphate dehydrogenase. Western blotting of CD63 under non-denaturing conditions. The two lanes are loaded with the same volume (15 μl) of samples. f Volcano plots showing that among differential expressed proteins, 43 upregulated and 133 downregulated in MVs from EBV positive GC tissues relative to EBV negative tissues. g Heatmap showing the quantification of differentially expressed proteins. Upregulated proteins are listed in the enlarged panel. Results from 3 biological replicates. h KEGG pathway analysis for the 176-differential expressed MV proteins. Top 10 KEGG pathways are shown based on enrichment score. i Diagram shows the screening strategies for key MV proteins specific to EBV infection. Representative of two independent experiments (a, d, e).
We then performed 4D-label free quantitative proteomics analysis for the isolated MVs, which identified a total of 4, 206 proteins; among them, 133 proteins showed significant downregulation and 43 proteins showed significant upregulation with >1.5 fold of change in MVs derived from EBV+ GC when compared to those from EBV− GC (Fig. 1f, g and Supplementary Data 1). KEGG analysis of these differentially regulated proteins (133 decreased and 43 increased) enriched pathways related to focal adhesion, T cell differentiation, Salmonella, EBV and other pathogen infection (Fig. 1h). 14 proteins were dramatically upregulated with >10 fold of change, 6 of which are related to GC according to The Cancer Genome Atlas (TCGA) database; and OLFM4 showed the highest abundance by proteomics analysis (Fig. 1i and Supplementary Data 2).
EBV infection induces OLFM4 expression and MV-mediated secretion
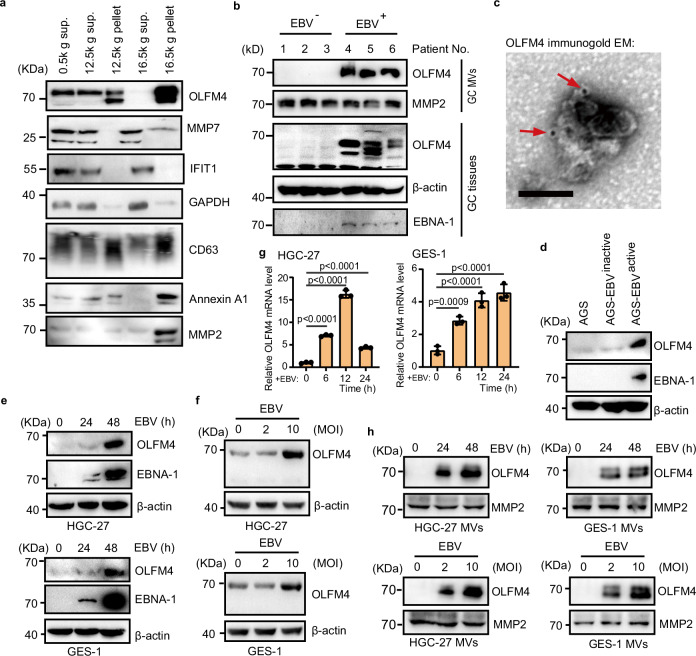
To validate the proteomics data of MVs, we examined by western blotting the expression levels of OLFM4, MMP7, and IFIT1 in various fractions isolated from human GC tissues using differential centrifugation. The OLFM4 protein was found to be significantly enriched in MVs (16,500 g pellet fraction), but we failed to detect IFIT1 in MVs (Fig. 2a and Supplementary Data 2). MMP7 can be detected in MVs, but mainly exists in the 16,500 g supernatant fraction that contains exosomes (Fig. 2a). It was reported that some virus such as EBV, Mayaro virus can induce the expression of metalloproteinase (MMP) proteins42–44, and the MMP7 protein level in serum-derived exosomes have been proposed as a diagnostic biomarker for GC45,46. Therefore, we focused on OLFM4 for subsequent investigation.
Fig. 2. OLFM4 is induced by EBV infection and secreted via MVs.
a MVs isolation from human gastric cancer tissues. Centrifugation fractions from each step are lysed for western blot. Lanes 1–4 are each loaded with 15 μl of sample. Lane 5 is loaded with 2 × 108 particles. b Immunoblotting showing OLFM4 expression in tumor tissues and their MVs of EBV-positive and EBV-negative GC patients. All lanes were loaded with 2 × 108 particles of total MV. c Representative TEM image of gastric cancer derived MV, immunogold-labeled with anti-OLFM4 antibodies. Arrowheads indicate 10-nm gold particles. Scale bar, 100 nm. d Immunoblotting analysis showing the OLFM4 levels in the indicated cells. AGS-EBV cells are treated with 0.5% rabbit anti-human immunoglobulin G (IgG) to activate EBV for 48 h. AGS cells are used as negative control. AGS-EBV (latency I) is labeled as AGS-EBVinactive. e Immunoblotting analysis showing the protein levels of OLFM4 and EBNA-1 in the HGC-27 cells (upper) and GES-1 cells (lower). HGC-27 and GES-1 cells were treated with 10 MOI EBV for 0, 24, and 48 h. f Immunoblotting showing the OLFM4 levels in the HGC-27 cells (upper) and GES-1 cells (lower). HGC-27 and GES-1 cells were treated with 0, 2 and 10 MOI of EBV for 48 h. g mRNA levels of OLFM4 in the HGC-27 and GES-1 cells after infection with 10 MOI EBV for 0, 6, 12, and 24 h (n = 3 biological replicates/group). h Immunoblotting showing the OLFM4 levels in the purified MVs from HGC-27 or GES-1 cells that treated with EBV by dose gradient (0, 2 and 10 MOI) or time course (0, 24, 48 h) as in (d, e). All lanes are loaded with 50 μg of total protein (d–f). Representative of two independent experiments (a–h). Data are presented as mean ± s.d., analyzed for significant differences by performing two-tailed, one-way ANOVA with Dunnett’s post hoc analysis (g).
Next, we detected OLFM4 protein level in tumor tissues and corresponding MVs of EBV-positive and EBV-negative GCs by western blotting. The protein level of OLFM4 was found to be significantly up-regulated in EBV-positive GC tissues, and the protein level of secreted OLFM4 was also found to be increased in MVs derived from the EBV-positive GCs (Fig. 2b), results indicating a positive correlation between OLFM4 protein levels in tissue and those in MVs. Further immune electron microscopy analysis showed that OLFM4 was localized on the membrane of MVs (Fig. 2c and Supplementary Fig. 2a).
To test the hypothesis that EBV infection induces expression and secretion of OLFM4 in GC, we used an EBV-infected GC cell line AGS-EBV (latency I), which was in an inactive state without producing EBV but could be activated to generate intact EBV once stimulated by Human-IgG. Our western blotting of EBNA-1 confirmed that only AGS-EBVactive cells produced EBVs while AGS and AGS-EBVinactive cells did not; and as expected, the cellular protein level of OLFM4 was dramatically increased upon active replication of EBV (Fig. 2d). In keeping with these results, co-culture of AGS cells derived from a GC cell line with medium differentiation state with Akata EBV+ cells derived from a Burkitt lymphoma cell line stably, also dose-dependently induced OLFM4 expression (Supplementary Fig. 2b).
Next, we treated GC cell line HGC-27 or gastric epithelial cell line GES-1 with EBV, the infection rate is about 20% (Supplementary Fig. 2c–g), and found that the protein expression of OLFM4 was significantly upregulated in a time- and dose-dependent manner (Fig. 2e, f). Further qPCR analysis in HGC-27 or GES-1 cells revealed at the mRNA level a significant upregulation of OLFM4 upon EBV infection (Fig. 2g and Supplementary Fig. 2h). Consistently, the protein expression of OLFM4 in MVs derived from HGC-27 or GES-1 cells was also found to be significantly increased upon EBV infection of the cells, indicating that EBV infection of a cell may alter the protein components of MVs derived from this cell (Fig. 2h). Moreover, we found that the expression of OLFM4 could also be induced by vesicular stomatitis virus (VSV) and herpes simplex virus type 1 (HSV-1) (Supplementary Fig. 2i–k). Together, these data demonstrate that EBV infection promotes the expression of OLFM4, leading to increased secretion of OLFM4 by MVs.
OLFM4 is a direct target gene of the cGAS-STING pathway
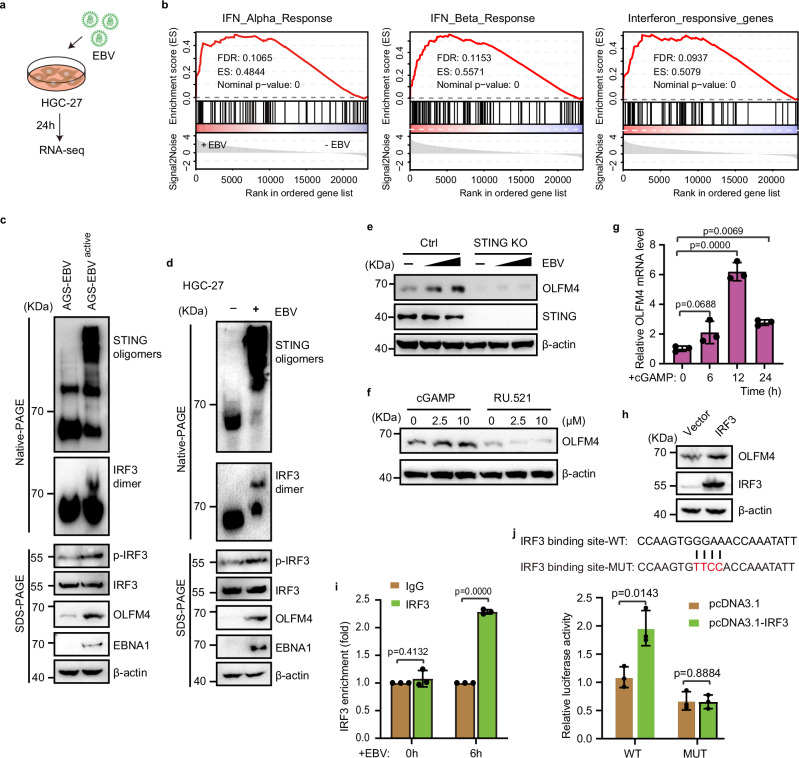
Given our finding that the mRNA level of OLFM4 was upregulated in EBV-infected cells, we next investigated how EBV regulates the transcription of OLFM4. To this end, we treated HGC-27 cells with EBV for 24 h, and then harvested the cells for RNA extraction and sequencing (Fig. 3a and Supplementary Data 3). It is well established that virus infection can activates the cGAS-STING signaling pathway to induce type I interferon response47. Indeed, Gene set enrichment analysis (GSEA) of the RNA sequencing data showed that the type I interferon (IFN-I) was significantly upregulated upon EBV infection (Fig. 3b). Consistent with this result, our western blot analysis in either AGS-EBVactive or EBV-treated HGC-27 cells revealed strong signals for STING oligomerization and IRF3 phosphorylation and dimerization, clearly evidencing activation of the cGAS-STING signaling pathway upon EBV infection (Fig. 3c, d).
Fig. 3. EBV infection induced OLFM4 by cGAS-STING pathway.
a Workflow for transcriptome analysis of EBV infected HGC-27 cells. b GSEA analysis for the enrichment of IFNα, IFNβ and interferon responsive genes in 10 MOI EBV infected HGC-27 cells (n = 3 replicates/group). c Immunoblotting showing STING signaling pathway effector protein activity in AGS-EBV-infected HGC-27 cells. d Immunoblotting showing the indicated proteins in EBV-infected HGC-27 cells. e Immunoblotting showing OLFM4 expression in STING-knockout AGS cells after EBV treatment. Cells were treated with 0, 5, 10 MOI EBV for 48 h. f Immunoblotting showing OLFM4 expression in HGC-27 cells treated with STING agonist (cGAMP) or cGAS inhibitor (RU.521). Cells were treated with cGAMP or RU.521 for 24 h. g mRNA levels of OLFM4 in cGAMP (10 μM)-treated HGC-27 cells. h Immunoblotting showing OLFM4 expression in HGC-27 cells transfected with pcDNA3.1-IRF3 plasmid (n = 3 biological replicates/group). i CHIP-qPCR analysis of the IRF3 binding activity on the promoter region (−278 to −258) of OLFM4 (n = 3 biological replicates/group). HGC-27 cells were treated with 10 MOI EBV for 0, 6 h. j Dual-luciferase reporter assay to detect the relative activity of OLFM4 −278 to −258 promotor region (n = 3 biological replicates/group). All lanes are loaded with 50 μg of total protein (c–f, h). Representative of two independent experiments (c–j). Data are presented as mean ± s.d., analyzed for significant differences by performing two-tailed, one-way ANOVA with Dunnett’s post hoc analysis (g, i, j).
To verify whether EBV-induced upregulation of OLFM4 is dependent on the cGAS-STING pathway, we knocked out STING in AGS cells and found that EBV infection was no longer able to induce the expression of OLFM4 in these cells (Fig. 3e). Consistent with this result, treating HGC-27 cells with the STING agonist cGAMP strongly promoted OLFM4 expression at both protein and mRNA levels, while treatment with the cGAS antagonist RU.521 decreased OLFM4 expression (Fig. 3f, g). Meanwhile, we found that HSV-1 treatment dose-dependently induced the upregulation of IFNB transcription in HGC-27 cells (Supplementary Fig. 3). At the same time, we observed that HSV-1 infection promotes OLFM4 expression in a dose-dependent manner (Supplementary Fig. 3), suggesting a positive correlation of OLFM4 transcription with the activation of cGAS-STING pathway.
To further explore whether IRF3, the major downstream transcription factor of the cGAS-STING pathway19, directly regulates the transcription of OLFM4 as a target gene, we analyzed the promoter region of OLFM4 by JASPAR database48, and found that the sequence of (-278)-CCAAGTGGGAAACCAAATATT-(-258) represents a potential IRF3-binding element. Indeed, overexpression of IRF3 in HGC-27 cells increased the expression of OLFM4 (Fig. 3h). Importantly, our chromatin immunoprecipitation with quantitative PCR (ChIP-qPCR) assay confirmed that IRF3 could bind to the promoter of OLFM4 and such binding was significantly enhanced upon EBV infection (Fig. 3i). Further dual-luciferase reporter assay showed that IRF3 activated the transcription of the reporter gene with the wild type IRF3-binding motif corresponding to the −278 to −258 region of the OLFM4 promoter, but not the one with a mutant IRF3-binding motif that would disrupt IRF3 binding (Fig. 3j). Together, these data demonstrate that EBV infection induces OLFM4 expression via the cGAS-STING pathway and OLFM4 is a direct target gene of this pathway.
EBV infection is positively correlated with OLFM4 expression levels in proximal GCs
To assess the clinical relevance of EBV-induced OLFM4 expression in GC, we first performed fluorescence in situ hybridization (FISH) analysis using EBER-1 probe to detect the EBV infection status of 168 clinical GC samples (Fig. 4a). The FISH results indicated that EBV+ GCs account for about 14.9% of the cases, with 11.3% cases being mild or modest and 3.6% cases being severe (Fig. 4b). Among the EBV+ GCs, 64% cases were found to be cardia cancers, suggesting that proximal GC is predisposed to EBV infection (Fig. 4c), consistent with previous study4. After quality control of the 168 clinical GC tissue microarray analysis results by immunohistochemistry (IHC), 154 of them were used to detect the expression levels of OLFM4 (Fig. 4d). The IHC results revealed that 29% of the cardia GCs have high levels of OLFM4 expression, yet only 14.8% of non-cardia GCs were OLFM4 high, suggesting that proximal GC is predisposed to high expression of OLFM4 (Fig. 4e and Supplementary Data 4).
Fig. 4. OLFM4 high expression positively correlated with EBV infection in gastric cancer.
a Representative image of EBV fluorescence in situ hybridization (FISH) from 168 clinical GC samples. EBER-1 RNA is used as the detection marker for EBV infection. Scale bars, 100 μm. b Pie chart showing t EBV infection rate. −: Negative (≤1 EBER-1 signal point); +: positive (2–100 EBER-1 signal points); ++: strong positive (>100 EBER-1 signal points). c Pie chart showing the tumor site preference of EBV positive gastric cancers. d Representative images for OLFM4 staining on tissue microarray analysis as described above. Negative; +: weak positive; ++: positive; +++: strong positive. Weak positive and positive samples were considered as low OLFM4 level, strong positive samples were considered as high OLFM4 level. Scale bars, 100 μm. e Pie chart showing OLFM4 expression status in cardia and non-cardia gastric tumors respectively. f Immunoblotting showing the OLFM4 levels in cardia and non-cardia gastric tumors. g Bar plots showing the proportion of strong-positive OLFM4 in different EBV infectious samples. 153 samples are verified by both EBV FISH and TMA analysis. h Correlation of OLFM4 expression with EBV infection by performing Chi-Squared test. i Immunoblotting showing the expression of OLFM4 and EBNA-1 in GC tumor samples. j Diagram showing the anatomic location distribution of EBV infection and OLFM4 high expression. All lanes are loaded with 50 μg of total protein (f, i). Representative of two independent experiments (f, i).
To further verify the correlation of high OLFM4 expression with cardia GCs, we extracted proteins from both cardia and non-cardia GC tissues for western blot, which showed that cardia GCs indeed have high OLFM4 expression more frequently than non-cardia GCs (Fig. 4f and Supplementary Fig. 4a–d). Further combinatorial analysis revealed a significant positive correlation between OLFM4 high expression and EBV infection, that is, EBVaGCs most likely (>80%) have high levels of OLFM4 expression (Fig. 4g, h). In keeping with these results, our western blotting analysis of a group of GC tissue samples also revealed a significant positive correlation between expression levels of EBNA-1 (as an indicator for EBV infection) and OLFM4 (Fig. 4i).
Because MVs are through to be circulated in the blood, we then collected EDTA-anticoagulated plasma and tissues samples from 7 EBV-positive and 10 EBV-negative GC patients. After isolation and characterization of MVs from plasma, we determined the protein levels of OLFM4 and found that OLFM4 was detectable in plasma MVs from almost all (6/7) EBV-positive GC patients (Supplementary Fig. 4e). However, OLFML4 protein was only detectable in MVs from 3 out of 10 EBV-negative GC patients (Supplementary Fig. 4e). Importantly, OLFM4 levels were somewhat increased in the MVs isolated from plasma and tissues of EBV-positive GC patients when compared to those of EBV-negative GC patients (Supplementary Fig. 4f). Together, these data indicate that EBV infection and OLFM4 expression are positively correlated with each other, both tend to occur in proximal or cardiac GCs (Fig. 4j).
OLFM4-containing MVs promote GC cell proliferation and tumor growth
HEK293T cells have been widely used to produce EVs due to their inherent rapid proliferation, high EV yield, ease of genetic manipulation49–52, low toxicity, and minimal immune response49,53. To assess the pathological role of MV-carried OLFM4, we overexpressed OLFM4 in HEK293T cells using pCDNA3.1 vector and collected MVs secreted by these cells. Meanwhile, MVs derived from HEK293T cells transfected with empty vector were used as controls. We first characterized the isolated MVs by TEM and nanoparticle tracking analysis (Supplementary Fig. 5a, b), and confirmed the expression of OLFM4 in MVs by western blotting (Supplementary Fig. 5c), mass spectrometry (Supplementary Data 5, 6), and Immuno-gold TEM (Supplementary Fig. 5d). Moreover, we found that OLFM4 was N-linked glycosylated and formed high-order oligomers in MVs (Supplementary Fig. 5e, f). In addition, we found almost no change of OLFM4 level in MVs when the secretion of exosomes was blocked by N-SMase inhibitor GW4869, confirming MV-specific secretion of OLFM4 (Supplementary Fig. 5g).
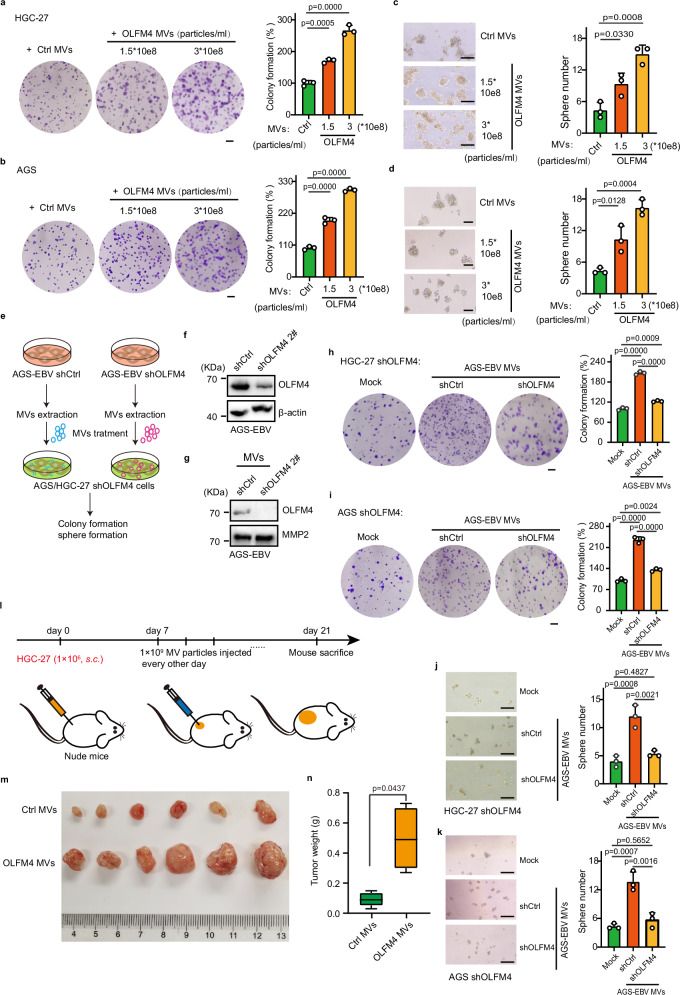
We then used the isolated MVs to treat cells and animals. Our colony formation assay showed that MVs carrying high levels of OLFM4 significantly increased the clone numbers (Fig. 5a, b) and sphere formation of HGC-27 and AGS cells (Fig. 5c, d). More importantly, we used the AGS–EBV cell model, a gastric epithelial cell line with stable EBV infection, to generate control and OLFM4-knockdown MVs by shRNA; and then used these MVs to treat HGC-27 and AGS cells (Fig. 5e–g). Note that the endogenous OLFM4 was also knocked down by shRNA in the recipient HGC-27 and AGS cells to avoid interference of OLFM4-MVs produced by these recipient cells themselves (Supplementary Fig. 5h). Consistent with the results of OLFM4-overexpressing MVs, the OLFM4-knockdown MVs were found to be unable to promote colony and sphere formation of the recipient cells as efficient as wildtype MVs (Fig. 5h–k).
Fig. 5. OLFM4 over expressing MV promotes gastric cancer progression.
a Colony formation in HGC-27 cells treated with OLFM4-overexpressing MVs (n = 3 biological replicates/group). Scale bars, 1.5 mm. b Colony formation in AGS cells treated with OLFM4-overexpressing MVs (n = 3 biological replicates/group). Scale bars, 1.5 mm. c Sphere formation in HGC-27 cells after treatment with OLFM4-overexpressing MVs (3 replicates/group). Scale bars, 200 μm. d Sphere formation in AGS cells treated with the indicated MVs (n = 3 biological replicates/group). Scale bars, 200 μm. e Experimental workflow. f Immunoblotting showing intracellular OLFM4 levels in OLFM4-knockdown AGS-EBV cells. Scramble shRNA is used as a control. g Immunoblotting showing the levels of OLFM4 in MVs derived from OLFM4-knockdown AGS-EBV cells. Each lane is loaded 2 × 108 MVs particles. h Colony formation of OLFM4-knockdown HGC-27 cells treated with the indicated MVs (n = 3 biological replicates/group). MVs are purified from shCtrl or shOLFM4-transfected AGS-EBV cells. Scale bars, 1.5 mm. i Colony formation ability of OLFM4-knockdown AGS cells treated with the indicated MVs (n = 3 biological replicates/group). Scale bars, 1.5 mm. j Sphere formation ability in OLFM4-knockdown HGC-27 cells after treatment with the indicated MV (n = 3 biological replicates/group). Scale bars, 200 μm. Sphere formation ability in OLFM4-knockdown AGS (k) cells after treatment with the indicated MV (n = 3 biological replicates/group). Scale bars, 200 μm. l Schematic illustration of subcutaneous tumorigenesis model and intratumoral MV injection in nude mice. m Representative images showing the tumor formation in HGC-27 after injection with OLFM4-expressing MV (n = 6 mice/group). n Box plot showing tumor weight in the indicated group. The center line corresponds to the median and box corresponds to the interquartile range. Representative of two independent experiments (a–j). Data are presented as mean ± s.d., analyzed for significant differences by performing two-tailed, unpaired Student’s t tests (n) or one-way ANOVA with Dunnett’s post hoc analysis (a–d, h–j).
Next, we seek to distinguish the role of MV-carried OLFM4 from that of intracellular OLFM4 in promoting cancer cell proliferation. To this end, we first assessed the effect of OLFM4 as a whole (both intracellular and secreted forms) on GC growth and found that knockdown of OLFM4 significantly decreased the colony and sphere formation of HGC-27 and AGS cells (Supplementary Fig. 5h–j). To better explore the secreted OLFM4 in a loss-of-function manner, we constructed a signal peptide deletion mutant of OLFM4, termed as OLFM4 (DelSP), which lacks the N-terminal 1-20 amino acid residues. We then transfected AGS or HGC-27 cells with wildtype and DelSP mutant OLFM4 and confirmed by western blotting that the DelSP mutant could not be secreted (Supplementary Fig. 5k, l). The results showed that overexpression of wildtype OLFM4, but not OLFM4 (DelSP) significantly promoted colony and sphere formation of HGC-27 and AGS cells (Supplementary Fig. 5m, n), indicating the functional importance of the secreted form of OLFM4.
To further validate the functional importance of the secreted form of OLFM4, we reconstituted OLFM4-knockdown HGC-27 cells with wildtype or DelSP mutant OLFM4 to either restore both intracellular or secreted function of OLFM4 or only restore its intracellular function. We confirmed the expression of OLFM4 in cell lysate and its absence in MVs in the case of the DelSP mutant (Supplementary Fig. 5o). Consistent with the above results showing functional importance of MV-carried OLFM4, OLFM4-knockdown HGC-27 cells reconstituted with DelSP mutant OLFM4 were found to have less viability than those reconstituted with wildtype OLFM4 (Supplementary Fig. 5p). Moreover, OLFM4-knockdown HGC-27 cells reconstituted with DelSP mutant OLFM4 receiving treatment with MVs generated by HEK293T cells expressing DelSP OLFM4 were found to have less viability than those receiving treatment with MVs generated by HEK293T cells expressing wildtype OLFM4 (Supplementary Fig. 5p). These results suggest that EBV-infected GC cells secrete OLFM4 MVs to stimulate overgrowth of neighboring cells (Supplementary Fig. 5q).
Subsequently, we used a xenograft tumor model to assess the biological function of MV-carried OLFM4 in vivo. To this end, 1 × 106 HGC-27 cells were injected into nude mice subcutaneously; after 1 week, 3 × 106 MVs particles with or without OLFM4 overexpression were injected into the grown tumors every other day; and the mice were sacrificed on day 21 for tumor measurement (Fig. 5l). Consistent with the results of colony and sphere formation assays, the size and weight of tumors receiving OLFM-overexpressed MVs were significantly larger and heavier than the control group (Fig. 5m, n). In addition, we assessed the function of OLFM4 MVs in tumor-bearing nude mice by intravenous injection; and found that OLFM4 MVs increased tumor weight and promoted tumor progression but not as significantly as intratumoral injection (Supplementary Fig. 5r).
OLFM4 acts as an extracellular inhibitor for Hippo signaling to promote YAP activation
To explore the mechanism through which MV-carried OLFM4 promotes tumor growth, we treated HGC-27 cells with OLFM4-overexpressing MVs and performed RNA sequencing (Fig. 6a). The results of RNA-seq identified 439 differentially expressed genes (p < 0.05) upon treatment with OLFM4-overexpressing MVs (Fig. 6b and Supplementary Data 7). Supporting this, a gene set enrichment analysis (GSEA) showed significant enrichment of YAP conserved signature genes in HGC-27 cells treated with OLFM4-overexpressing MVs (Fig. 6c). A subsequent real-time qPCR assay confirmed that MV-carried OLFM4 significantly upregulated the mRNA expression of YAP target genes CTGF and CYR61 in both HGC-27 cells (Fig. 6d) and HGC-27 cells-derived tumors (Supplementary Fig. 6a, b).
Fig. 6. OLFM4 inhibits Hippo signal pathway.
a Schematic illustration of transcriptome analysis in HGC-27 cells after treatment with OLFM4-overexpressing MVs (n = 3 biological replicates/group). b Volcano plot showing 208 up-regulated genes and 231 down-regulated genes (p < 0.05) in HGC-27 cells treated with OLFM4-overexpressing MVs (n = 3 biological replicates/group). c GSEA plots showing the distribution of YAP signature genes identified from OLFM4 over expressing MVs treated HGC-27 cells. d mRNA levels of YAP target genes (CTGF, CYR61) in OLFM4 over expressing MVs treated HGC-27 cells (n = 3 biological replicates/group). e Immunoblotting showing the levels of p-MOB1, MOB1, p-YAP1 and YAP1 in HGC-27 cells treated with OLFM4-overexpressing MVs. f Immunoblotting showing the indicated proteins in OLFM4-overexpressing MV-treated AGS cells. g Immunoblotting showing the translocalization of YAP in HGC-27 cells after treatment with the indicated MVs. Histone and GAPDH are used as the markers for nuclear and cytoplasm, respectively. Nuc. nucleus, Cyt. Cytoplasm. h Immunofluorescence analysis of YAP and OLFM4 in HGC-27 cells (n = 3 wells/group). Quantification for YAP nuclear translocation is shown. Scale bars, 10 μm. All lanes were loaded with 50 μg of total protein (e, f). Representative of two independent experiments (d–h). Data are presented as mean ± s.d., analyzed for significant differences by performing two-tailed, unpaired Student’s t tests (d, h).
Next, we examined Hippo-YAP signaling western blot and found that MVs isolated from HEK293T cells overexpressing OLFM4 significant inhibited the phosphorylation of both YAP (Ser127) and MOB1 (Thr35) in HGC-27 and AGS cells (Fig. 6e, f). These results were further confirmed by western blotting and immunofluorescence analyses showing strong nuclear translocation of YAP in HGC-27 cells treated with MV-carried OLFM4 (Fig. 6g, h). Similarly, MVs extracted from human GC tissues containing higher expression level of OLFM4 also showed inhibitory effect on Hippo signaling (Supplementary Fig. 6e). Moreover, overexpression of OLFM4 in HGC-27 cells also significantly inhibited the Hippo signaling in terms of phosphorylation of YAP and MOB1 (Supplementary Fig. 6d). Furthermore, purified OLFM4 protein alone was found also able to inhibit the Hippo signaling (Supplementary Fig. 6e). On the contrary, HGC-27 cells transfected with shOLFM4 showed increased phosphorylation levels of YAP and MOB1 (Supplementary Fig. 6f). We also assessed the effect of intracellular OLFM4 on Hippo-YAP signaling and found that OLFM4 (DelSP) significantly reduced the phosphorylation of both Mob1 and YAP1 in HGC-27 cells (Supplementary Fig. 6g) and increased the mRNA levels of YAP target genes (Supplementary Fig. 6h). Meanwhile, AGS cells treated with MMP7 MVs generated in HEK293T cells did not show significant change in Hippo signaling and colony and sphere formation (Supplementary Fig. 6i–p). Together, these results indicate that tumor cells can secrete and deliver OLFM4 via MVs as an extracellular inhibitor for the Hippo signaling in recipient cells.
MV-carried OLFM4 inhibits the Hippo signaling of recipient cells via binding FAT1
Finally, we investigated the molecular mechanism by which OLFM4 inhibits the Hippo signaling and activates YAP in recipient cells. Since MVs can be internalized by target cells through fusion and uptake54, we first examined the localization of OLFM4 in recipient cells. To this end, we isolated MVs from HEK293T cells overexpressing HA-tagged OLFM4, and treated HGC-27 cells with these MVs. Both immunofluorescence and western blot results revealed that MV-delivered OLFM4 was localized on the plasma membrane of the recipient cells (Fig. 7a, b), suggesting that OLFM4 acts on the plasma membrane to inhibit the Hippo signaling. Considering that OLFM4 can bind pan-cadherin35 and that FAT1 is a nonclassical cadherin protein which previously identified as a key upstream regulator of the Hippo pathway28,55, we speculated that OLFM4 may regulate the activity of upstream Hippo signaling through FAT1. To test this hypothesis, we assessed the interaction of OLFM4 with FAT1. Our immunofluorescence assay confirmed that MV-delivered HA-tagged OLFM4 was well colocalized with FAT1 on recipient cells (Fig. 7c). Subsequent domain mapping and pulldown assays using purified recombinant proteins revealed that OLFM4 directly interacted with the cadherin repeats #28-33 of FAT1 (Fig. 7d, e). By contrast, such colocalization signal was undetectable in the case of truncated variants of FAT1, including residues 4023-4588 lacking the N-terminal cadherin repeats #28-33 or residues 3790-4588 lacking the cadherin repeats #28-33 and transmembrane sequence (Supplementary Fig. 7a). Moreover, intracellular region (residues 3790-4588) of FAT1 can’t interact with OLFM4 (Del SP) (Supplementary Fig. 7b). These observations further suggest that MV-carried OLFM4 interacts with the extracellular region of FAT1 and do so only when FAT1 is localized on the plasma surface.
Fig. 7. OLFM4 binds with FAT1 cadherin domain and block hippo signaling.
a Immunofluorescence detection of the localization of OLFM4-carried by MVs on recipient cells. AGS cells is treated with HEK293T derived HA-OLFM4 over expressing MVs (3 × 108 particles/ml) for 24 h. Wheat germ agglutinin (WGA) is used to label plasma membranes. Scale bars, 10 μm. b Immunoblotting showing the cellular localization of OLFM4-carried MV-treated AGS cells. Annexin A1 and GAPDH are used as markers for membrane and cytoplasm, respectively. Cyt. Cytoplasm, Mem. membrane. c Immunofluorescence detection showing the localization of FAT1 and OLFM4 in HGC-27 cells. Scale bars, 10 μm. d GST-pulldown assay showing the interaction between OLFM4 and #28-33 cadherin domain of FAT1. Extracellular cadherin motifs in FAT1 and GST-OLFM4 construction are shown. e Immunoprecipitation showing the interaction of OLFM4 with FAT1 cadherin 28-33. Construction of FLAG tagged FAT1 with cadherins #28-33, TM and ICD. f Immunoprecipitation showing the interaction between FLAG-tagged FAT1 and MST1 in HEK293FT cells with or without HA-OLFM4 transfection. Cells were treated for 2 h at 4 °C with DMSO or the reversible crosslinker DSP prior to cell lysis and immunoprecipitation. g Immunoblotting showing the expression of p-YAP and YAP in FAT1-knockdown HGC-27 cells treated with 0, 1.5, and 3 × 108 particles/ml HEK293T-derived OLFM4 overexpressing MVs. All lanes are loaded with 50 μg of total protein. Knockdown efficiency of FAT1 is shown by qPCR assay (n = 3 biological replicates/group). Data are presented as mean ± s.d., analyzed for significant differences by performing two-tailed, unpaired Student’s t tests. h Scheme depicting the proposed molecular mechanism model. Representative of two independent experiments (a–g).
Previous studies have demonstrated that FAT1 assembles via its intracellular domain a multi protein signaling complex including the Hippo kinases MST1/2, LATS1/2 and adapter protein MOB1 on the plasma membrane to facilitate the activation of MST1/229. To test whether OLFM4 binding to FAT1 would interfere with this complex, we performed Co-IP in HEK293T cells and found that OLFM4 significantly inhibited the interaction of FAT1 with MST1, but not LATS1 and MOB1 (Fig. 7f). Moreover, knockdown of FAT1 significantly blocked the inhibitory effect of MV-carried OLFM4 on the Hippo signaling (Fig. 7g). Together, these data indicate that MV-carried OLFM4 acts as a secreted inhibitor for the Hippo signaling through a mechanism in which OLFM4 binds to extracellular domain of FAT1 to disrupt the interaction of FAT1 intracellular domain with MST1, thereby limiting MST1 activation (Fig. 7h).
Discussion
Both viral infection and extracellular vesicles have been extensively implicated in tumorigenesis, yet the specific mechanisms remain obscure. With the improved understanding of gastric cancer, a subset of gastric cancer patients infected with EBV has been identified. Here we report OLFM4, a well-recognized intestinal stem cell marker, as an MV-carried Hippo inhibitor that is clinically associated with EBVaGC.
Previously, OLFM4 has been reported as a molecular marker for intestinal stem cells and some gastrointestinal cancers37,39,40,56. OLFM4 has been implicated in cancer cell proliferation, cell cycle regulation, and cell adhesion and metastasis of some malignancies, especially in gastrointestinal cancers37,39. It has been reported that higher expression of OLFM4 is associated with severity of disease for many viral and bacterial infections57. In this work, we showed that EBV and other types of virus (VSV, HSV-1) can led to significant upregulation of OLFM4. Given the positive relationship between the cGAS-STING activation and OLFM4 expression, we speculate that in other virus-associated tumors, OLFM4 expression is induced with a context of cGAS-STING activation.
Early studies have shown that OLFM4 promotes GC cell migration and proliferation58, and reduced expression of OLFM4 in GC is associated with lymph node and distant metastases and with poor prognosis59. Partially consistent with these observations, here we found that OLFM4 MVs act as an extracellular inhibitory ligand for the Hippo pathway—it is possible that OLFM4 MVs may also regulate GC metastasis via Hippo or Hippo-related pathways. Upstream of OLFM4 MVs, we identified OLFM4 as a direct target gene of the cGAS-STING signaling pathway. Upon EBV infection, activated cGAS-STING promotes OLFM4 expression via IRF3. Notably, we observed that EBV infection significantly also stimulated intracellular MMP7 expression and its MVs secretion. However, MMP7 MVs did seem to have any effect on the Hippo pathway and GC growth. Given the previous report that YAP signaling upregulates MMP7 expression and promote cell adhesion and migration60–62, we speculate that OLFM4 MVs activate YAP signaling, which may in turn stimulate MMP7 expression and secretion.
Dysregulation of Hippo pathway leads to aberrant cell growth and a variety of diseases, including cancer. Multiple membrane proteins including GPCRs and FAT1 have been identified as upstream regulators of the Hippo signaling pathway26,27. However, the possible extracellular ligands for these regulators remain to be identified. In the current study, we identified OLFM4 as a MV-carried secretory inhibitor for the Hippo signaling in recipient cells. FAT1 has been reported as an important factor causing dysregulation of the Hippo pathway29. Here, we show that MV-carried OLFM4 mainly anchors on the MV surface and transferred onto the plasma surface of recipient cells during the fusion of MVs to the recipient cells, and that these surface-anchored OLFM4 only interacts with membrane FAT1 but not cytosol FAT1 to inhibit the interaction with MST1, thereby downregulating Hippo signaling in recipient cells (Supplementary Fig. 7c). This work exemplifies from a perspective of cell-cell communication and signal transduction cascade how tumor cells reshape the microenvironment, which not only provides insights into the mechanisms underlying upstream in particular extracellular signaling of the Hippo pathway, but also explains how Hippo dysregulation contributes to tumorigenesis in EBVaGC and other types of cancers with viral infection.
Limitations of this study
MVs in the tumor microenvironment could target multiple types of cells including normal epithelial cells, immune cells and tumor cells. In this study, we mainly investigated tumor cells as target cells, but the full spectrum of the recipient cell types of OLFM4-containing MVs is not defined. It has been reported that YAP activation in peritumoral cells can inhibit tumors by cell competition, and the relative activation level of the YAP/TAZ in tumor cells and peritumoral cells determines whether the tumor grows63. In this regard, our study showed that once being infected with virus, tumor cells can release MV-carried OLFM4 to inhibit in a paracrine manner the Hippo signaling in neighboring cells (Supplementary Fig. 7), which may in turn interfere the cell-cell competition in the tumor microenvironment. In the future, how OLFM4 acts as an extracellular Hippo inhibitor to regulate YAP-mediated cell-cell competition during tumorigenesis warrants further investigation.
Additionally, HEK293T cells have been widely used as EV producer cells due to their inherent rapid proliferation, high EV yield, and ease of genetic manipulation49–52. The stem cells or metabolic active cells are thought to internalize significantly more HEK293T EVs than terminally differentiated cells49. Theoretically, as most EVs under evaluation are derived from human cells, they may elicit immune responses in mice. However, some studies have assessed the immune response and toxicity in mice, and found that HEK293T EVs showed low toxicity, and minimal changes in immune markers49,53. Our study demonstrate a regulatory mechanism though which viral infection is coupled via MVs with intercellular control of the Hippo signaling. Although our mass spectrometry analysis on HEK293T (with and without OLFM4 overexpression) MVs, and found these MVs did not contain T-antigen (Supplementary Data 5 and 6), we do not fully elucidate whether MVs from EBV-infected cell or HEK293T cell may elicit immune response in recipient cells. Further investigations are warranted to address this issue and better define the toxicity and immune response of these used MVs.
Methods
Ethics statement
The study was conducted in compliance with the guidelines of the Institutional Animal Care and Use Committee of Shanghai Tenth People’s Hospital (approval ID, SHDSYY-2023-P0011).
Gastric cancer specimen collection
Gastric cancer tissues were obtained from patients with gastric cancer who underwent gastrectomy as described previously64. Data on clinicopathological features and prognoses of the patients were collected and analyzed retrospectively. Patients were signed informed consents for the use of the specimen. The study was performed in accordance with the Declaration of Helsinki and approved by the Huashan Hospital Institutional Review Board, Fudan University (approval ID, No.2017-222).
Cell lines and expression of EBV
The HEK293T, HGC-27, GES-1 and AGS cells were purchased from American Type Culture Collection (ATCC). EBV-infected cell lines AGS-EBV (latency I) and Akata were kindly provided by Professor Chun-kui Shao (Sun Yat-sen University). HEK293T cells were cultured in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. HGC-27, AGS, GES-1, AGS-EBV, and Akata cells were cultured in RPMI1640 medium (Gibco, USA) supplemented with 10% FBS and 1% penicillin-streptomycin. All cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C. Epstein-Barr virus was produced as previous described65. In brief, 5 × 106 EBV-positive Akata cells were suspended in 5 ml medium containing 0.5% (v/v) Rabbit anti-human immunoglobulin G (IgG) (GeneTex, USA) and incubated for 2 h at 37 °C, then washed and resuspended and incubated in fresh RPMI medium for 48 h. The culture medium was harvested and filtered through a 0.45 μm-pore-size membrane and stored at −80 °C for further experiments.
Generation of cell lines
To knock down OLFM4 in GC cell lines, short hairpin RNAs (shRNAs) against human OLFM4 (target 1: AGTGCAGAGCATTAACTATAA, target 2: GAGGGAATATGTCCAATTAAT), or scrambled shRNA control were constructed in pLKO.1, and then co-transfected with the viral packaging plasmids psPAX and pMD2.G to package lentiviral particles using HEK293T cells. After 48–72 h transfection, lentiviral supernatants were filtered through a 0.45 μm-pore-size membrane and stored at −80 °C. For generate a stable OLFM4 knock down cell line, HGC-27 cells were infected with the lentivirus in fresh culture media containing 8 μg/mL polybrene overnight. Then change cells to fresh media 48 h after infection, and cells were selected by 1 μg/ml puromycin for about 1 week. STING Knockout procedure and primers was used as described by Hayman et al.66.
Antibodies and regents
Antibodies against Annexin A1, OLFM4, GAPDH, MST1, MOB1, pMOB1(T35), LATS1, Histone H2B, pYAP(S127), HA tag, IRF3, p-IRF3, and STING were purchased from Cell Signaling Technology (USA). The CD63, YAP, EBNA-1 and FAT1 mouse monoclonal antibodies were purchased from Santa Cruz (USA). Antibodies anti-MMP2, MMP7, and β-actin was from abcam (Shanghai, China). antibodies against FLAG tag, and 6xHis tag antibody was purchased from Proteintech (USA). The HRP-conjugated secondary antibodies including goat anti-rabbit goat anti-mouse were purchased from Thermo Fisher Scientific (USA). CHIP grade IRF3 antibody, Alexa Fluor 488 conjugated Goat anti-mouse and Alexa Fluor 568 conjugated Goat anti-Rabbit secondary antibodies were from Thermo Fisher Scientific (USA). The detailed information and diluted ratio of antibodies are listed in Supplementary Data 8.
Exosome N-SMase inhibitor GW4869 was purchased from Selleck (USA). cGAMP was purchased from MedChemExpress. Rabbit anti-human immunoglobulin G (IgG) and RU.521 was purchased from Selleck (USA). Dual-Lumi™ Luciferase Assay Kit was from Beyotime (China). Wheat Germ Agglutinin (WGA) and DSP cross-linker were purchased from Thermo Fisher Scientific (USA). HSV-1 (KOS) virus was purchased from BrainVTA (Wu Han, China). The detailed information of reagents is listed in Supplementary Data 8.
Plasmids and transfection
To construct 6xHis tagged FAT1, the FAT1 cadherin #1-9 (1-1139 aa)/cadherin #10-18 (1140-2081 aa)/cadherin #19-27 (2082-3023 aa)/cadherin #28-33 (3024-3789 aa) were cloned in pET28a vector to express recombinant FAT1 proteins in Escherichia coli strain BL21 (DE3). The 3xFLAG tagged FAT1 cadherin #28-33 (3024-3789 aa) with transmembrane domain (TM) and intracellular domain (ICD) (3790-4588 aa) was cloned into pCDN3.1(+) vector for cell expression, the 3×FLAG tag was inserted after signal peptide (1-21 aa) of FAT1. Wild type OLFM4, or OLFM4 deleted of the signal peptide (DelSP) (1-20 aa) were cloned into pCDNA3.1(+) vector. HA or 3×FLAG tag was inserted after the signal peptide (1-20 aa) of OLFM4 and cloned into pCDN3.1(+) vector. shRNAs were cloned into pLKO.1 vector, sgRNA was cloned in lentiCRISPR v2 plasmid.
Immunoprecipitation
FLAG-FAT1 and HA-OLFM4 plasmids were co-transfection into HEK293T cells, after 48 h, cells were washed with ice cold PBS and treated with DSP cross-linker according to the instructions, then cells were lysed in NP40 buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 10 mM NaF, 1 mM sodium orthovanadate, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF) and Protein inhibitor cocktail (MedChemExpress)). After centrifugation, 20 µL of anti-FLAG M2 beads (Merk, USA) were added in the lysate and incubated on a rocker for 4 h at 4 °C. Beads were washed 3 times with NP40 buffer. Then the immunoprecipitate was subjected to western blot. For HA-OLFM4 immunoprecipitation, HEK293T cells were transfected with pCDNA3.1(+)-HA-OLFM4 vector. After 48 h incubation, cells were lysed in NP40 buffer, and the supernatant were incubated with HA beads for 4 h at 4 °C. Beads were washed 3 times with NP40 buffer.
Western blot analysis
Cells or purified microvesicles were lysed on ice in RIPA buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 0.1% SDS, 5 mM EDTA, 2 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 1% Triton X-100, 10 mM NaF, 0.5 mM DTT, 1 mM PMSF, Protease Inhibitor Cocktail), Supernatants containing the solubilized proteins were quantified by BCA Protein Assay Kit. After boiled for 10 min, equal amounts of samples by mass were separated using 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with 5% defatted milk at room temperature for 1 h, and incubated with the corresponding primary antibodies at recommended dilutions overnight at 4 °C. After washed with PBS, membranes were incubation with HRP-conjugated secondary antibodies at room temperature for 1 h. The blots on the membranes were developed with ECL detection reagents (Yeasen Biotechnology, China).
For Native PAGE, cells were lysed in lysis buffer (150 mM NaCl, 1% NP-40,50 mM Tris-HCl (PH7.4), 1 mM PMSF, and Protease inhibitor cocktail) with a protease inhibitor cocktail. The samples were centrifuged to remove insoluble precipitates. The protein concentration was measured using BCA Protein Assay Kit. The gel was pre-run for 30 min, then protein was mixed with loading buffer without SDS and subjected to native gel electrophoresis, Electrophoresis was performed on ice under low voltage.
RNA extraction and quantitative PCR (qPCR)
RNA was extracted from treated cells or tissues using RNA isolator (Vazyme, China) following manufacturer’s instructions. One microgram of total RNA from each sample was converted to cDNA with HiScript® II 1st Strand cDNA Synthesis Kit (Vazyme, China). For the qPCR reactions, SYBR green master mix (Vazyme, China) was used. The primers were designed in PrimerBank67. GAPDH was used as an internal control. Human OLFM4 (Primer F: ACTGTCCGAATTGACATCATGG, Primer R: TTCTGAGCTTCCACCAAAACTC), human GAPDH (Primer F: GGAGCGAGATCCCTCCAAAAT, Primer R: GGCTGTTGTCATACTTCTCATGG), human CTGF (Primer F: ACCGACTGGAAGACACGTTTG, Primer R: CCAGGTCAGCTTCGCAAGG), human CYR61 (Primer F: CTCGCCTTAGTCGTCACCC, Primer R: CGCCGAAGTTGCATTCCAG).
RNA sequencing
RNA from cells were extracted using the RNA isolater Reagent (Vazyme, China) according to the manufacturer’s protocol. After quantification and qualification, a total amount of 3 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (NEB, E7775) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. The library preparations were sequenced on Illumina Novaseq 6000. FPKM (Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced) of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analysis was performed using the DESeq2 R package. The p values were adjusted using the Benjamini and Hochberg method. Corrected p value of 0.05 and absolute fold-change of 2 were set as the threshold for significantly differential expression.
Tissue microarray, immunohistochemical staining and fluorescence in situ hybridization (FISH)
The gastric cancer and normal tissue microarray sections were prepared by Shanghai Outdo Biotech (Shanghai, China). The tissue array contains 141 cases of gastric tumors with paired normal tissue, and 28 cases of gastric tumor without paired normal tissue. Tissue arrays were used for OLFM4 IHC staining analysis. The same TMA sections were used for EBV FISH experiments, EBER-1 probe (5’-CTCCTCCCTAGCAAAACCCAGGACGGCC-3’) was used to detect EBV infected cells in gastric cancer samples.
Protein purification and GST pulldown
OLFM4 without signal peptide (1-20 aa) was cloned into pGEX4T-2. GST-OLFM4 protein was expressed in BL21 (DE3) by 0.5 M IPTG inducing overnight, then harvest cells and resuspend in phosphate-buffered saline (PBS) (0.1% Triton100, Protease Inhibitor Cocktail, 1 mM PMSF), after lyzed by sonication, proteins were purified by glutathione Sepharose 4B (GE healthcare). The GST-OLFM4 protein was eluted with 300 μl 10 mM reduced glutathione (in 50 mM Tris). The FAT1 extracellular cadherin domains were arranged into four fragments (Cadherin 1-9, 10-18, 19-27, and 28-33), each fragment was cloned into pET28a with N-terminal 6xHis tag. His tagged FAT1 proteins were expressed in BL21(DE3) by 0.5 M IPTG inducing overnight, cells were collected and disrupted by sonication in lysis buffer (20 mM Tris, 500 mM KCl, 10% glycerol, 10 mM imidazole, 1 mM PMSF). After 12,000 × g centrifugation for 30 min, cell lysates were incubated with Ni-NTA agarose beads for 4 h, then wash the beads with wash buffer (20 mM imidazole in lysis buffer), finally elute proteins by elution buffer (300 mM imidazole in lysis buffer).
For pull down assay, GST-tagged OLFM4 and 6xHis-tagged FAT1 fragments were mixed in PBS containing 0.5% Triton X-100 and 1 mM PMSF, then pre-blocked glutathione Sepharose 4B beads was added and incubated for 4 h at 4 °C. Twenty µg of each proteins were added in the reaction. After washing three times with PBS, proteins were eluted from beads with 10 mM Glutathione. The eluent was subjected to SDS-PAGE, and proteins were detected by immunoblot.
Tissue derived microvesicles isolation
Surgical samples were rinsed in cold sterile PBS solution, and then thoroughly minced using fine scissors in a volume of PBS as described by Mayr et al.68. The enrichment of MVs is carried out by differential centrifugation as described previously69. In brief, Supernatants were centrifuged at 500 × g for 10 min, then 12,500 × g for 5 min to remove cell debris and dead cells. Microvesicles were pelleted after centrifugation at 16,500 × g for 45 min and resuspended in PBS. MVs were quantified by BCA assays and were stored at −80 °C for further analysis.
Cell line derived microvesicles isolation
To purification microvesicles from cell culture supernatants, cells were cultured in media supplemented with 10% FBS which was filtered by a 0.22 μm pore size filter. Supernatants were collected from 48 h cell cultures, the culture supernatants were centrifuged at 500 × g for 10 min, then 12,500 × g for 5 min to remove cell debris and dead cells. Microvesicles were pelleted after centrifugation at 16,500 × g for 45 min and resuspended in PBS as described69. Then MVs were stored at −80 °C for further analysis.
Transmission electron microscopy
The transmission electron microscopy assays were performed as previously described69. In brief, MVs derived from human gastric tissue or HEK293T cell lines were suspended in PBS, then dropped on formvar carbon-coated copper grids. After 1 min, the grid was washed with PBS for 3 times, then negatively stained with 2% uranyl acetate for 1 min, staining was removed with a paper filter, and grids were air-dried and visualized using a Tecnai G2 Spirit 120 KV (FEI) transmission electron microscope.
For immunogold labeling, purified MVs were suspended in PBS, then placed on formvar carbon-coated copper grids. Blocked with 1% BSA for 20 min, and incubated with Rabbit anti OLFM4 antibody overnight, followed by incubation with the goat anti-Rabbit secondary antibody conjugated with 10 nm gold particles (Sigma-Aldrich) for 2 h at room temperature. Each staining step was followed by five PBS washes and ten ddH2O washes before staining with 2% uranyl acetate.
Nanoparticle tracking analysis
The nanoparticle tracking analysis assays were carried out as previously described69. In brief, the nanoparticle concentration and size distribution of MVs purified from cell culture supernatants or gastric cancer tissues were measured using a NanoSight NS300 (Malvern), which is equipped with a Blue488 nm laser and a high sensitivity sCMOS camera. During measurements, temperature was set and kept constant at 25 °C. The averaged value for each biological replicate was used to determine the mode of the size distribution and the particle concentration of MV.
Immunofluorescence staining
For immunofluorescence, cells were seeded on the Glass Bottom Cell Culture Dish (NEST). After treatment with MV, the cells were washed with ice-cold PBS for 3 times, and fixed with 4% paraformaldehyde in PBS. Then the cells were permeabilized with 0.1% Triton X-100 in PBS, and blocked with 3% of bovine serum albumin (BSA) in PBS for 1 h. Fixed cells were incubated with primary antibodies overnight at 4 °C. Wash the cells with PBS 3 times, then incubate them with fluorophore-conjugated secondary antibodies for 1 h. Nuclei were stained with DAPI. Images were observed using Zeiss LSM900 microscope equipped at ×63 magnification (Carl Zeiss).
Mass spectrometry
Gastric cancer tissues (3 EBV-positive, 3 EBV-negative) were used for MVs isolation (3 biological, two technical replication). Proteins extracted from MVs were quantified by BCA Protein Assay Kit (Pierce, USA). Then reduced by TCEP (tris (2-carboxyethyl) phosphine) for 60 min at 37 °C. Following alkylation by add IAM (Iodoacetamide) for 40 min at room temperature under dark conditions. After Trypsin digestion, peptides were desalted and analyzed by an EASY nLC-1200 system (Thermo, USA) coupled with a timsTOF Pro2 (Bruker, Germany) mass spectrometer at Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Briefly, the C18-reversed phase column (75 μm ×25 cm, Ionopticks, USA) was equilibrated by solvent A (2% ACN with 0.1% formic acid). The peptides were eluted using the following gradient: 0–45 min, 3%–28% solvent B (80% ACN with 0.1% formic acid); 45–50 min, 28%–44% solvent B; 50–55 min, 44%–90% solvent B; 55–60 min, 90%–90% solvent B. The tryptic peptides were separated at a flow rate of 250 nl/min. The electrospray voltage was 1.5 kV. The secondary MS scanning range was 100–1700 m/z. Data acquisition on the timsTOF Pro2 was collected using the parallel accumulation serial fragmentation (PASEF) acquisition mode. After the first MS stage, the second MS stage (charge number of the parent ions was 0–5) was recorded using the 10 PASEF mode. A dynamic exclusion time of 24 s was used for the MS/MS scan.
MS/MS spectra were searched using MaxQuant version 2.0.3.1 software70 against uniprot_taxonomy_9606_unique.fasta database. The highest score for a given peptide mass (best match to that predicted in the database) was used to identify parent proteins. The parameters for protein searching were set as follows: tryptic digestion with upto two missed cleavages, carbamidomethylation of cysteines as fixed modification, and oxidation of methionines and protein N-terminal acetylation as variable modifications. False discovery rate (FDR) of peptide identification was set as FDR ≤ 0.01. A minimum of one unique peptide identification was used to support protein identification. The differential regulated proteins were analyzed by two tailed unpaired t-test.
Chromatin immunoprecipitation (ChIP)
HGC-27 cells treated with EBV for 0 and 6 h were cross linked by adding 37% formaldehyde for 15 min, then quenched by adding glycine to a final concentration of 125 mM for 5 min. After washed by cold PBS, CHIP lysis buffer (50 mM Hepes/KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton-X 100, 0.1% Na-deoxycholate, Protease inhibitor cocktail) was added to the cells. Scrape off cells then centrifuge samples and remove the supernatant, resuspend cells with 300 μL CHIP lysis buffer. Sonicate the lysis with a bioruptor, 420 W, 30 s on, 30 s off, 10 min. Spin the lysis at 10,000 × g for 5 min at 4 °C, CHIP grade IRF3 antibody (Thermo Fisher Scientific) was added to the supernatant and incubate overnight. Add pre-washed protein G-sepharose and incubate for 2 h, then wash the beads with lysis buffer, lysis buffer with 0.5 M NaCl, Tris/LiCl buffer (10 mM Tris, 0.25 M LiCl, 0.5% NP40, 0.5% Na-deoxycholate, 1 mM EDTA), and Tris/EDTA buffer (10 mM Tris, 0.5% NP-40, 0.5% Na-deoxycholate, 1 mM EDTA) for 5 min respectively. Directly add 100 μL 10% chelex (Biorad) to the washed beads, boil samples for 10 min. Add proteinase K and heat at 55 °C for 30 min. Boil samples for 10 min, spin down beads and collect supernatant, stored at −80 °C for further qPCR assays. qPCR primers for IRF3 binding site on OLFM4 (Primer F: AGACTACTTTGGTCTCATCTC, Primer R: CTGCCAAACATTAACTCAAATG).
Luciferase reporter assays
The luciferase reporter assays were performed using Dual Lumpi™ II Double Luciferase reporter gene detection kit (Beyotime) according to the manufacturer’s protocol. Briefly, the cells were transfected with appropriate plasmids in 24-well plates. After 48 h, cells were collected and lysed for luciferase assay. The relative luciferase activity was normalized with renilla luciferase activity. The promoter region of OLFM4 was amplified with the primers: GCTCAGGTACCTTCCTCCATGGAGCCTCCAAAC (Kpn1 site) and GCTCCAAGCTTCCACTCTGTGAGCTGCCCTTGG (HindIII site), and was subcloned into the pGL3 basic firefly luciferase reporter vector. The amplified PCR fragments were then used as a template for generating promoter construct carrying mutation using specific primers.
Cell viability assay
The cell death and viability assays are conducted as described by An et al.71. In brief, cells were seeded into 96-well plates at a density of 1000 per well overnight to have them become attached to the wells, and then treated with HEK293T-delivered MVs at indicated concentrations. The cells were incubated for the indicated length of time, and then used with an ATPbased CellTiter-LumiTM Plus kit (Beyotime) according to the manufacturer’s instructions. The intracellular ATP contents were measured using a BioTek Synergy TM NEO multi-detector microplate reader (Thermo). Cell viability was calculated using the equation, % Cell viability = [value (test) − value (blank)] × [[value (control) − value (blank)]] −1 × 100.
Colony formation and tumor spheroid formation assay
For colony formation assays, 2000 cells were placed into each well of 6-cm dishes and cultured for 2 weeks. Then, the cells were fixed with methanol for 20 min and stained with 0.1% crystal violet in PBS for 15 min. The stained colonies were counted by ImageJ software.
Spheroid formation assays were performed in ultralow attachment 6-well plates (Corning, USA). Briefly, 8000 cells were seeded per well, and then cultured in RPMI1640 medium which added 50 ng/ml EGF and 20 ng/ml bFGF for 1 week.
Subcutaneous tumorigenesis
For establishing human gastric cancer model in nude mice, HGC-27 cells (1 × 106 cells in 100 μl medium) were injected into flanks of 4-week-old male Balb/c nude mice, with an inoculation volume of 0.1 ml. one week later, subcutaneous masses began to appear.
For Intratumoral injection, HEK293T derived OLFM4 over expressing microvesicles (3 × 108 particles in 10 μl PBS) began to be injected into the tumor every other day. Mice were euthanized 21 days after cell inoculation. Immediately following euthanasia, tumors were harvested for imaging and tumor weight were measured. The tumor tissues were then used for RNA extraction and RT qPCR analysis.
For intravenous MVs administration. Once tumors reached a size of 100 mm3, mice were injected through the lateral tail vein with HEK293T derived Ctrl or OLFM4 over expressing MVs (1 × 109 MV particles in 100 μl HEPES buffer) every other day. All mice were euthanized 21 days after cell inoculation. The maximal tumor burden permitted by the ethics committee is no more than 2000 mm3. Tumors were harvested for imaging and tumor weight were measured.
Statistical analysis
Both cellular and animal studies tended to be underpowered. Estimation of sample size for planned comparisons of two independent means using a two-tailed test were undertaken using the GraphPad Prism 9. Data are presented as mean ± s.d. for continuous variables and as frequencies and proportions for categorical variables. Continuous data were compared using Student’s t test (comparing two variables) or one-way ANOVA with Dunnett’s post hoc analysis (comparing multiple variables). Categorical variables were analyzed using Chi-square test. A value of p < 0.05 was considered to indicate a significant difference. At least 2 biological replicates were used throughout the study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
Our work was supported by National Key R&D Program of China (2020YFA0803200, 2023YFC2505903), the National Natural Science Foundation of China (31930026, 32200567, 92168116, 82373251, 82222052, 82372613, 82150112, 82361168638, 32070710, 32270747, 82072740, 32170706), the Science and Technology Commission of Shanghai Municipality (23YF1432900, 22ZR1448100, 22QA1407200, 22QA1407300, 23ZR1480400, 23ZR1448900), the Guiding Funds of Central Government for Supporting the Development of the Local Science and Technology of Shanxi Province (YDZJSX2021B016, YDZJSX2021B015), China Postdoctoral Science Foundation (2019M651610) and Shanghai Super Postdoctoral Incentive Program. We thank Chun-kui Shao (from Sun Yat-sen University) for providing AGS-EBV and Akata cell lines.
Author contributions
F.W. performed most of cellular experiments. Y.H. and Zhang.Z. did animal assay. Hui.Z. and F.C. performed immunofluorescence and TMA experiments. W.W. and W.Q. constructed plasmids and biochemical assay. L.A. performed ChIP experiments. J.J., Y.M., J.Y., Y.T., F.L., Huan.Z. and Y.Z. discussed and analyzed the results. S.J., Z.Zhou and F.W. designed the experiments and wrote the manuscript. S.J., Zhao.Z., Y.X. and W.B. supervised the project. All authors read and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks Jianmin Zhang and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE240276. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository72,73 with the dataset identifier PXD057977. Source data are provided with this paper.
Competing interests
The authors have filed patents (CN2024116656372) regarding the potential diagnostic application of OLFM4-containing MVs.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fuping Wen, Yi Han, Hui Zhang, Zhangting Zhao, Wenjia Wang.
Contributor Information
Wenqi Bai, Email: wenqibai2022@163.com.
Yuanzhi Xu, Email: xuyuanzhi@tongji.edu.cn.
Zhaocai Zhou, Email: zhouzhaocai@fudan.edu.cn.
Shi Jiao, Email: jiaoshi@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-54850-6.
References
- 1.Dunmire, S. K., Verghese, P. S. & Balfour, H. H. Jr. Primary Epstein-Barr virus infection. J. Clin. Virol.102, 84–92 (2018). [DOI] [PubMed] [Google Scholar]
- 2.AbuSalah, M. A. H. et al. Recent advances in diagnostic approaches for Epstein-Barr virus. Pathogens9, 10.3390/pathogens9030226 (2020). [DOI] [PMC free article] [PubMed]
- 3.Imai, S., Nishikawa, J. & Takada, K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol.72, 4371–4378 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corvalan, A. et al. Epstein-Barr virus in gastric carcinoma is associated with location in the cardia and with a diffuse histology: a study in one area of Chile. Int. J. Cancer94, 527–530 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature513, 202–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeppesen, D. K. et al. Reassessment of exosome composition. Cell177, 428–445 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andaloussi, S. E. L., Mager, I., Breakefield, X. O. & Wood, M. J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov.12, 347–357 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Fu, M. et al. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol. Cancer18, 41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, T. et al. The roles of extracellular vesicles in gastric cancer development, microenvironment, anti-cancer drug resistance, and therapy. Mol. Cancer18, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maia, J., Caja, S., Strano Moraes, M. C., Couto, N. & Costa-Silva, B. Exosome-based cell-cell communication in the tumor microenvironment. Front. Cell Dev. Biol.6, 18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rojas, A., Araya, P., Gonzalez, I. & Morales, E. Gastric tumor microenvironment. Adv. Exp. Med. Biol.1226, 23–35 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Hu, C. et al. Exosome-related tumor microenvironment. J. Cancer9, 3084–3092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wortzel, I., Dror, S., Kenific, C. M. & Lyden, D. Exosome-mediated metastasis: communication from a distance. Dev. Cell49, 347–360 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Yang, J., Liu, Z., Zeng, B., Hu, G. & Gan, R. Epstein-Barr virus-associated gastric cancer: a distinct subtype. Cancer Lett.495, 191–199 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Saito, M. & Kono, K. Landscape of EBV-positive gastric cancer. Gastric Cancer24, 983–989 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Meckes, D. G. Jr. et al. Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl Acad. Sci. USA107, 20370–20375 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meckes, D. G. Jr. Exosomal communication goes viral. J. Virol.89, 5200–5203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmisano, G. et al. Characterization of membrane-shed microvesicles from cytokine-stimulated beta-cells using proteomics strategies. Mol. Cell Proteom.11, 230–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet.20, 657–674 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Wang, Y. et al. cGAS-STING pathway in cancer biotherapy. Mol. Cancer19, 10.1186/s12943-020-01247-w (2020). [DOI] [PMC free article] [PubMed]
- 21.Zhou, R. et al. The triggers of the cGAS-STING pathway and the connection with inflammatory and autoimmune diseases. Infect. Genet. Evol.77, 10.1016/j.meegid.2019.104094 (2020). [DOI] [PubMed]
- 22.Zheng, Y. & Pan, D. The Hippo signaling pathway in development and disease. Dev. Cell50, 264–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao, Z., An, L., Han, Y., Jiao, S. & Zhou, Z. The Hippo signaling pathway in gastric cancer. Acta Biochim. Biophys. Sin.10.3724/abbs.2023038 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, J. et al. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell122, 421–434 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Meng, Z., Moroishi, T. & Guan, K. L. Mechanisms of Hippo pathway regulation. Genes Dev.30, 1–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, Y. et al. Transmembrane protein KIRREL1 regulates Hippo signaling via a feedback loop and represents a therapeutic target in YAP/TAZ-active cancers. Cell Rep.40, 10.1016/j.celrep.2022.111296 (2022). [DOI] [PubMed]
- 27.Yu, F. X. et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell150, 780–791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed, A. F. et al. FAT1 cadherin acts upstream of Hippo signalling through TAZ to regulate neuronal differentiation. Cell Mol. Life Sci.72, 4653–4669 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, D. et al. Assembly and activation of the Hippo signalome by FAT1 tumor suppressor. Nat. Commun.9, 2372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastushenko, I. et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature589, 448–455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bu, J. et al. KK-LC-1 as a therapeutic target to eliminate ALDH+ stem cells in triple negative breast cancer. Nat. Commun.14, 10.1038/s41467-023-38097-1 (2023). [DOI] [PMC free article] [PubMed]
- 32.Xu, J. et al. Secreted stromal protein ISLR promotes intestinal regeneration by suppressing epithelial Hippo signaling. EMBO J.39, e103255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi, Q., Li, D. Y., Luo, H. R., Guan, K. L. & Ye, K. Netrin-1 exerts oncogenic activities through enhancing Yes-associated protein stability. Proc. Natl Acad. Sci. USA112, 7255–7260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, J. et al. Identification and characterization of a novel member of olfactomedin-related protein family, hGC-1, expressed during myeloid lineage development. Gene283, 83–93 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Liu, W., Chen, L., Zhu, J. & Rodgers, G. P. The glycoprotein hGC-1 binds to cadherin and lectins. Exp. Cell Res.312, 1785–1797 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi, D., Koshida, S., Moriai, R., Tsuji, N. & Watanabe, N. Olfactomedin 4 promotes S-phase transition in proliferation of pancreatic cancer cells. Cancer Sci.98, 334–340 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guette, C., Valo, I., Vétillard, A. & Coqueret, O. Olfactomedin-4 is a candidate biomarker of solid gastric, colorectal, pancreatic, head and neck, and prostate cancers. Proteomics Clin. Appl.9, 58–63 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Liu, W. & Rodgers, G. P. Olfactomedin 4 expression and functions in innate immunity, inflammation, and cancer. Cancer Metastasis Rev.35, 201–212 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Yu, L., Wang, L. & Chen, S. Olfactomedin 4, a novel marker for the differentiation and progression of gastrointestinal cancers. Neoplasma58, 9–13 (2011). [DOI] [PubMed] [Google Scholar]
- 40.van der Flier, L. G., Haegebarth, A., Stange, D. E., van de Wetering, M. & Clevers, H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology137, 15–17 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Kawasaki, K. et al. LGR5 induces β-catenin activation and augments tumour progression by activating STAT3 in human intrahepatic cholangiocarcinoma. Liver Int.41, 865–881 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Bengue, M. et al. Mayaro virus infects human chondrocytes and induces the expression of arthritis-related genes associated with joint degradation. Viruses11, 10.3390/v11090797 (2019). [DOI] [PMC free article] [PubMed]
- 43.Real, F. et al. S100A8-mediated metabolic adaptation controls HIV-1 persistence in macrophages in vivo. Nat. Commun.13, 5956 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan, Y. Y. et al. Epstein-Barr virus Zta upregulates matrix metalloproteinases 3 and 9 that synergistically promote cell invasion in vitro. PLoS ONE8, e56121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanco-Calvo, M. et al. Circulating levels of GDF15, MMP7 and miR-200c as a poor prognostic signature in gastric cancer. Future Oncol.10, 1187–1202 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Liu, L. et al. A novel strategy to identify candidate diagnostic and prognostic biomarkers for gastric cancer. Cancer Cell Int.21, 335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan, D., Jiang, W. & Hao, J. Research advances in how the cGAS-STING pathway controls the cellular inflammatory response. Front. Immunol.11, 615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castro-Mondragon, J. et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res.50, D165–D173 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jurgielewicz, B. J., Yao, Y. & Stice, S. L. Kinetics and specificity of HEK293T extracellular vesicle uptake using imaging flow cytometry. Nanoscale Res. Lett.15, 170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferguson, S., Kim, S., Lee, C., Deci, M. & Nguyen, J. The phenotypic effects of exosomes secreted from distinct cellular sources: a comparative study based on miRNA composition. AAPS J.20, 67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnsen, K. B. et al. A comprehensive overview of exosomes as drug delivery vehicles—endogenous nanocarriers for targeted cancer therapy. Biochim. Biophys. Acta1846, 75–87 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Faruqu, F. N., Xu, L. & Al-Jamal, K. T. Preparation of exosomes for siRNA delivery to cancer cells. J. Vis. Exp. 10.3791/58814 (2018). [DOI] [PMC free article] [PubMed]
- 53.Zhu, X. et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J. Extracell. Vesicles6, 1324730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meldolesi, J. Exosomes and ectosomes in intercellular communication. Curr. Biol.28, R435–R444 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Li, Z. et al. Loss of the FAT1 tumor suppressor promotes resistance to CDK4/6 inhibitors via the Hippo pathway. Cancer Cell34, 893–905.e898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jang, B. G. et al. Distribution of intestinal stem cell markers in colorectal precancerous lesions. Histopathology68, 567–577 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Liu, W. & Rodgers, G. P. Olfactomedin 4 is a biomarker for the severity of infectious diseases. Open Forum Infect. Dis. 9, 10.1093/ofid/ofac061 (2022). [DOI] [PMC free article] [PubMed]
- 58.Ran, X. et al. A quantitative proteomics study on olfactomedin 4 in the development of gastric cancer. Int. J. Oncol.47, 1932–1944 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Luo, Z. et al. OLFM4 is associated with lymph node metastasis and poor prognosis in patients with gastric cancer. J. Cancer Res. Clin. Oncol.137, 1713–1720 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Yuan, J. Q., Zhang, K. J., Wang, S. M. & Guo, L. YAP1/MMP7/CXCL16 axis affects efficacy of neoadjuvant chemotherapy via tumor environment immunosuppression in triple-negative breast cancer. Gland Surg.10, 2799–2814 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Du, F. et al. Sex determining region Y-box 12 (SOX12) promotes gastric cancer metastasis by upregulating MMP7 and IGF1. Cancer Lett.452, 103–118 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Nukuda, A. et al. Stiff substrates increase YAP-signaling-mediated matrix metalloproteinase-7 expression. Oncogenesis4, e165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moya, I. M. et al. Peritumoral activation of the Hippo pathway effectors YAP and TAZ suppresses liver cancer in mice. Science366, 1029–1034 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Tang, Y. et al. Selective inhibition of STRN3-containing PP2A phosphatase restores Hippo tumor-suppressor activity in gastric cancer. Cancer Cell38, 115–128.e119 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Kassis, J. et al. EBV-expressing AGS gastric carcinoma cell sublines present increased motility and invasiveness. Int. J. Cancer99, 644–651 (2002). [DOI] [PubMed] [Google Scholar]
- 66.Hayman, T. J. et al. STING enhances cell death through regulation of reactive oxygen species and DNA damage. Nat. Commun.12, 2327 (2021). [DOI] [PMC free article] [PubMed]
- 67.Wang, X., Spandidos, A., Wang, H. & Seed, B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res.40, D1144–D1149 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayr, M. et al. Proteomics, metabolomics, and immunomics on microparticles derived from human atherosclerotic plaques. Circ. Cardiovasc. Genet.2, 379–388 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Chen, G. et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature560, 382–386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinitcyn, P. et al. MaxQuant goes Linux. Nat. Methods15, 401 (2018). [DOI] [PubMed] [Google Scholar]
- 71.An, L. et al. Combinatorial targeting of Hippo-STRIPAK and PARP elicits synthetic lethality in gastrointestinal cancers. J. Clin. Invest.132, 10.1172/jci155468 (2022). [DOI] [PMC free article] [PubMed]
- 72.Chen, T. et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res.50, D1522–D1527 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res.47, D1211–D1217 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE240276. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository72,73 with the dataset identifier PXD057977. Source data are provided with this paper.