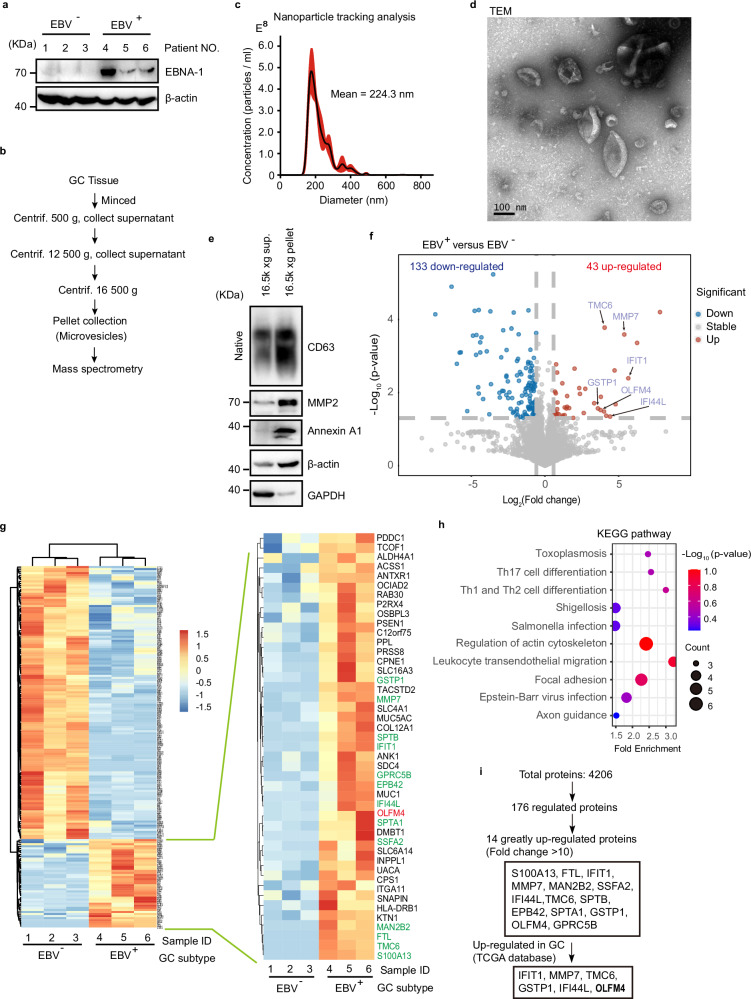
Fig. 1. Quantitative proteome study of MVs from EBVaGC.
a Western blotting to validate EBV infection of GC tissues used for MV extraction. EBV+ and EBV− human GC tumor samples are used for protein extraction and western blot. EBNA-1, Epstein-Barr virus (EBV)-encoded nuclear antigen 1, a marker protein of EBV. All lanes are loaded with 50 μg of total protein. b Centrifugation protocol and workflow for microvesicle enrichment from gastric cancer tissues. c Representative nanoparticle tracking analysis. d Transmission electron microscopy (TEM) imaging of MVs from gastric cancer tissues. Scale bar represents 100 nm. e Western blot of MVs isolated from gastric cancer for conventional extracellular vesicle markers. GAPDH glyceraldehyde-3-phosphate dehydrogenase. Western blotting of CD63 under non-denaturing conditions. The two lanes are loaded with the same volume (15 μl) of samples. f Volcano plots showing that among differential expressed proteins, 43 upregulated and 133 downregulated in MVs from EBV positive GC tissues relative to EBV negative tissues. g Heatmap showing the quantification of differentially expressed proteins. Upregulated proteins are listed in the enlarged panel. Results from 3 biological replicates. h KEGG pathway analysis for the 176-differential expressed MV proteins. Top 10 KEGG pathways are shown based on enrichment score. i Diagram shows the screening strategies for key MV proteins specific to EBV infection. Representative of two independent experiments (a, d, e).