Abstract
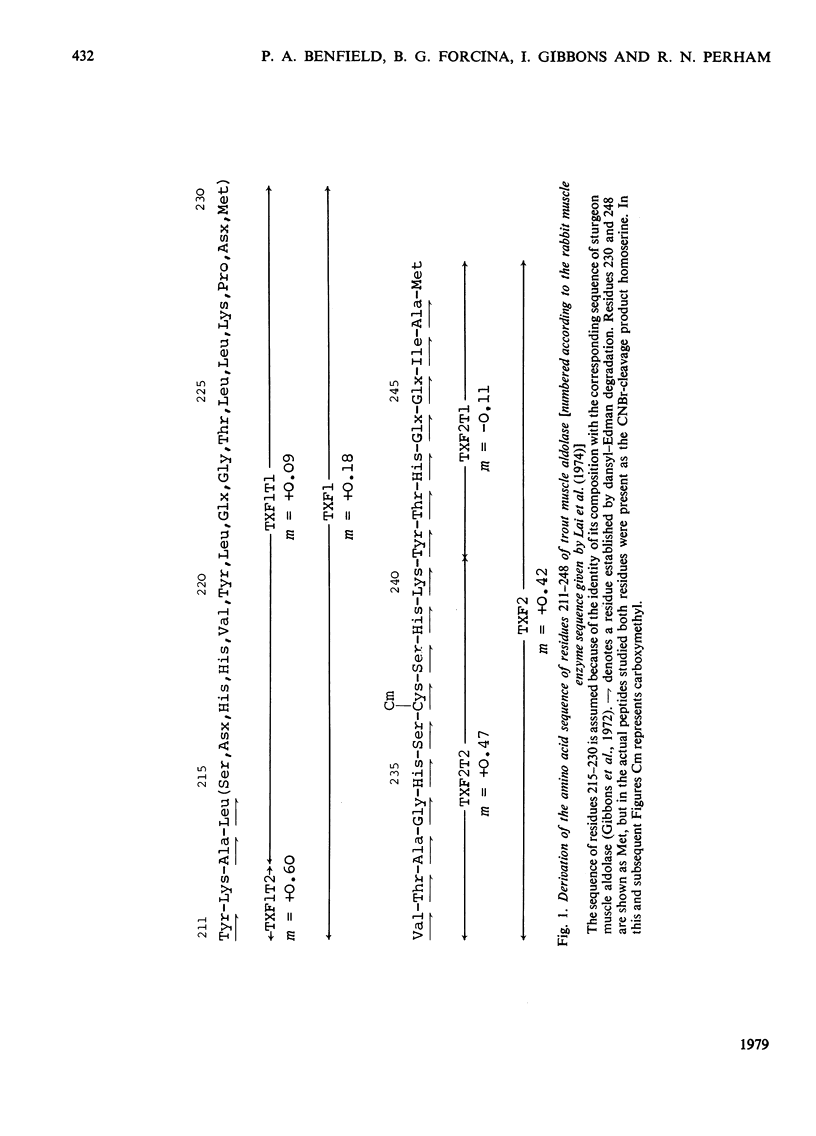
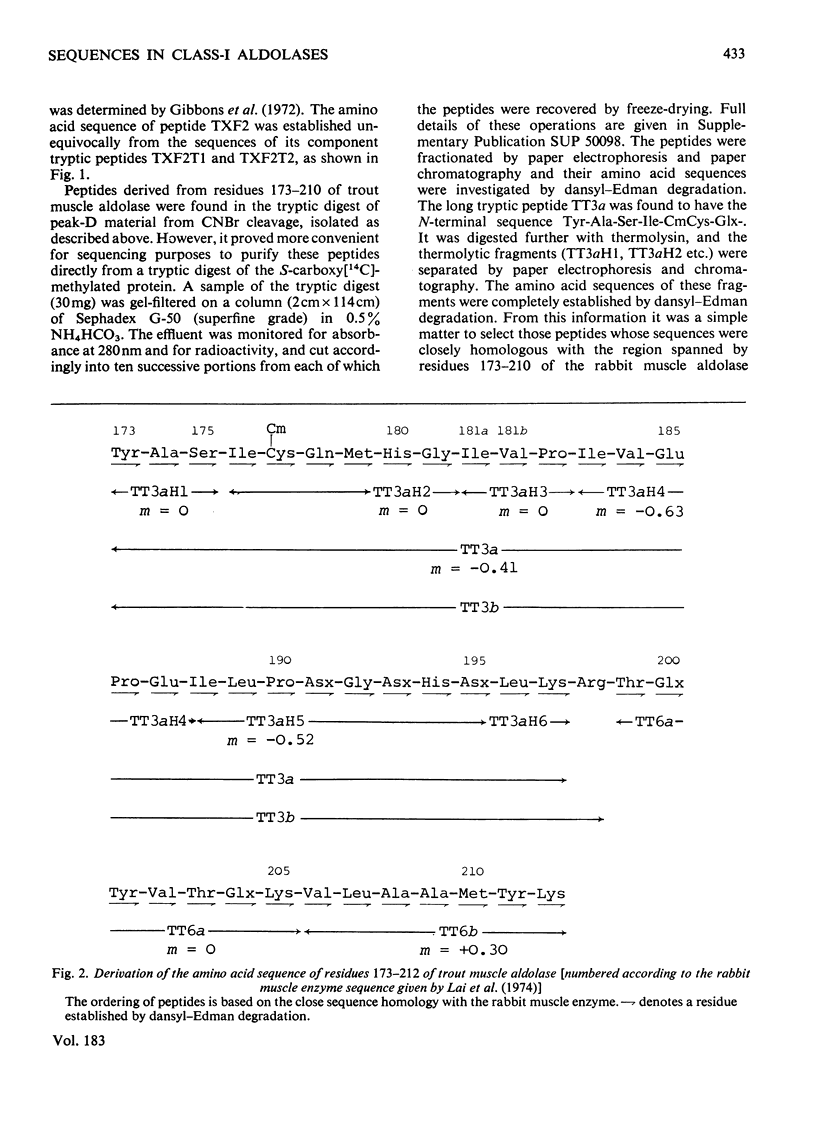
1. Amino acid sequences covering the region between residues 173 and 248 [adopting the numbering system proposed by Lai, Nakai & Chang (1974) Science 183, 1204-1206] were derived for trout (Salmo trutta) muscle aldolase and for ox liver aldolase. A comparable sequence was derived for residues 180-248 of sturgeon (Acipenser transmontanus) muscle aldolase. The close homology with the rabbit muscle enzyme was used to align the peptides of the other aldolases from which the sequences were derived. The results also allowed a partial sequence for the N-terminal 39 residues for the ox liver enzyme to be deduced. 2. In the light of the strong homology evinced for these enzymes, a re-investigation of the amino acid sequence of rabbit muscle aldolase between residues 181 and 185 was undertaken. This indicated the presence of a hitherto unsuspected -Ile-Val-sequence between residues 181 and 182 and the need to invert the sequence -Glu-Val- to -Val-Glx- at positions 184 and 185. 3. Comparison of the available amino acid sequences of these enzymes suggested an early evolutionary divergence of the genes for muscle and liver aldolases. It was also consistent with other evidence that the central region of the primary structure of these enzymes (which includes the active-site lysine-227) forms part of a conserved folding domain in the protein subunit. 4. Detailed evidence for the amino acid sequences proposed has been deposited as Suy Lending Division, Boston Spa, Wetherby, West Yorkshire LS23 7BQ, U.K., from whom copies can be obtained on the terms indicated in Biochem. J. (1978) 169, 5.
Full text
PDF













Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P. J., Gibbons I., Perham R. N. A comparative study of the structure of muscle fructose 1,6-diphosphate aldolases. Eur J Biochem. 1969 Dec;11(3):503–509. doi: 10.1111/j.1432-1033.1969.tb00802.x. [DOI] [PubMed] [Google Scholar]
- Anderson P. J., Gibson D. The amino acid sequence of the N-terminal region of aldolase. Can J Biochem. 1973 May;51(5):514–519. doi: 10.1139/o73-063. [DOI] [PubMed] [Google Scholar]
- Anderson P. J., Perham R. N. The reactivity of thiol groups and the subunit structure of aldolase. Biochem J. 1970 Apr;117(2):291–298. doi: 10.1042/bj1170291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLOSTEIN R., RUTTER W. J. COMPARATIVE STUDIES OF LIVER AND MUSCLE ALDOLASE. II. IMMUNOCHEMICAL AND CHROMATOGRAPHIC DIFFERENTIATION. J Biol Chem. 1963 Oct;238:3280–3285. [PubMed] [Google Scholar]
- Baldwin S. A., Perham R. N. Novel kinetic and structural properties of the class-I D-fructose 1,6-bisphosphate aldolase from Escherichia coli (Crookes' strain). Biochem J. 1978 Mar 1;169(3):643–652. doi: 10.1042/bj1690643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Iasio A., Trombetta G., Grazi E. Fructose 1,6-bisphosphate aldolase from liver: the absolute configuration of the intermediate carbinolamine. FEBS Lett. 1977 Feb 1;73(2):244–246. doi: 10.1016/0014-5793(77)80990-3. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. The structures of cytochrome c and the rates of molecular evolution. J Mol Evol. 1971;1(1):26–45. doi: 10.1007/BF01659392. [DOI] [PubMed] [Google Scholar]
- Dixon H. B., Perham R. N. Reversible blocking of amino groups with citraconic anhydride. Biochem J. 1968 Sep;109(2):312–314. doi: 10.1042/bj1090312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagles P. A., Johnson L. N., Joynson M. A., McMurray C. H., Gutfreund H. Subunit structure of aldolase: chemical and crystallographic evidence. J Mol Biol. 1969 Nov 14;45(3):533–544. doi: 10.1016/0022-2836(69)90310-6. [DOI] [PubMed] [Google Scholar]
- Forcina B. G., Perham R. N. Amino acid sequence homology between muscle and liver aldolases. FEBS Lett. 1971 Oct 15;18(1):59–63. doi: 10.1016/0014-5793(71)80406-4. [DOI] [PubMed] [Google Scholar]
- Gibbons I., Anderson P. J., Perham R. N. Amino acid sequence homology in the active site of rabbit and sturgeon muscle aldolases. FEBS Lett. 1970 Sep 18;10(1):49–53. doi: 10.1016/0014-5793(70)80413-6. [DOI] [PubMed] [Google Scholar]
- Gibbons I., Perham R. N., Anderson P. J. Amino-acid sequence homology in the muscle aldolases from sturgeons of different species. Nat New Biol. 1972 Aug 9;238(84):173–175. doi: 10.1038/newbio238173a0. [DOI] [PubMed] [Google Scholar]
- Gibbons I., Perham R. N. The reaction of aldolase with 2-methylmaleic anhydride. Biochem J. 1970 Mar;116(5):843–849. doi: 10.1042/bj1160843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORECKER B. L., ROWLEY P. T., GRAZI E., CHENG T., TCHOLA O. THE MECHANISM OF ACTION OF ALDOLASES. IV. LYSINE AS THE SUBSTRATE-BINDING SITE. Biochem Z. 1963;338:36–51. [PubMed] [Google Scholar]
- Heidner E. G., Weber B. H., Eisenberg D. Subunit structure of aldolase. Science. 1971 Feb 19;171(3972):677–679. doi: 10.1126/science.171.3972.677. [DOI] [PubMed] [Google Scholar]
- Lacko A. G., Brox L. W., Gracy R. W., Horecker B. L. The carboxyl-terminal structure of rabbit liver aldolase (aldolase B). J Biol Chem. 1970 Apr 25;245(8):2140–2141. [PubMed] [Google Scholar]
- Lai C. Y., Chen C., Horecker B. L. Primary structure of two COOH-terminal hexapeptides from rabbit muscle aldolase: a difference in the structure of the alpha and beta subunits. Biochem Biophys Res Commun. 1970 Jul 27;40(2):461–468. doi: 10.1016/0006-291x(70)91031-4. [DOI] [PubMed] [Google Scholar]
- Lai C. Y., Nakai N., Chang D. Amino acid sequence of rabbit muscle aldolase and the structure of the active center. Science. 1974 Mar;183(130):1204–1206. doi: 10.1126/science.183.4130.1204. [DOI] [PubMed] [Google Scholar]
- Lambert J. M., Perham R. N., Coggins J. R. Intramolecular ionic interactions of lysine residues and a possible folding domain in fructose diphosphate aldolase. Biochem J. 1977 Jan 1;161(1):63–71. doi: 10.1042/bj1610063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebherz H. G., Rutter W. J. A class I (Schiff base) fructose diphosphate aldolase of prokaryotic origin. Purification and properties of Micrococcus aerogenes aldolase. J Biol Chem. 1973 Mar 10;248(5):1650–1659. [PubMed] [Google Scholar]
- London J. Variations in the quaternary structure of three lactic acid bacteria aldolases. Evidence for the existence of a class I and class II aldolase in Lactobacillus casei. J Biol Chem. 1974 Dec 25;249(24):7977–7983. [PubMed] [Google Scholar]
- Midelfort C. F., Mehler A. H. Deamidation in vivo of an asparagine residue of rabbit muscle aldolase. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1816–1819. doi: 10.1073/pnas.69.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildvan A. S., Kobes R. D., Rutter W. J. Magnetic resonance studies of the role of the divalent cation in the mechanism of yeast aldolase. Biochemistry. 1971 Mar 30;10(7):1191–1204. doi: 10.1021/bi00783a016. [DOI] [PubMed] [Google Scholar]
- Offord R. E. Electrophoretic mobilities of peptides on paper and their use in the determination of amide groups. Nature. 1966 Aug 6;211(5049):591–593. doi: 10.1038/211591a0. [DOI] [PubMed] [Google Scholar]
- Penhoet E. E., Kochman M., Rutter W. J. Ioslation of fructose diphosphate aldolases A, B, and C. Biochemistry. 1969 Nov;8(11):4391–4395. doi: 10.1021/bi00839a025. [DOI] [PubMed] [Google Scholar]
- Perham R. N., Thomas J. O. The subunit molecular weights of the alpha-ketoacid dehydrogenase multienzyme complexes from E. coli. FEBS Lett. 1971 Jun 2;15(1):8–12. doi: 10.1016/0014-5793(71)80066-2. [DOI] [PubMed] [Google Scholar]
- RUTTER W. J. EVOLUTION OF ALDOLASE. Fed Proc. 1964 Nov-Dec;23:1248–1257. [PubMed] [Google Scholar]
- Sajgó M., Hajós G. The amino acid sequence of rabbit muscle aldolase. Acta Biochim Biophys Acad Sci Hung. 1974;9(3):239–241. [PubMed] [Google Scholar]
- Shapiro A. L., Maizel J. V., Jr Molecular weight estimation of polypeptides by SDS-polyacrylamide gel electrophoresis: further data concerning resolving power and general considerations. Anal Biochem. 1969 Jun;29(3):505–514. doi: 10.1016/0003-2697(69)90335-2. [DOI] [PubMed] [Google Scholar]
- Stellwagen E. Predicted structure for aldolase. J Mol Biol. 1976 Sep 25;106(3):903–911. doi: 10.1016/0022-2836(76)90273-4. [DOI] [PubMed] [Google Scholar]
- Stribling D., Perham R. N. Purification and characterization of two fructose diphosphate aldolases from Escherichia coli (Crookes' strain). Biochem J. 1973 Apr;131(4):833–841. doi: 10.1042/bj1310833. [DOI] [PMC free article] [PubMed] [Google Scholar]


