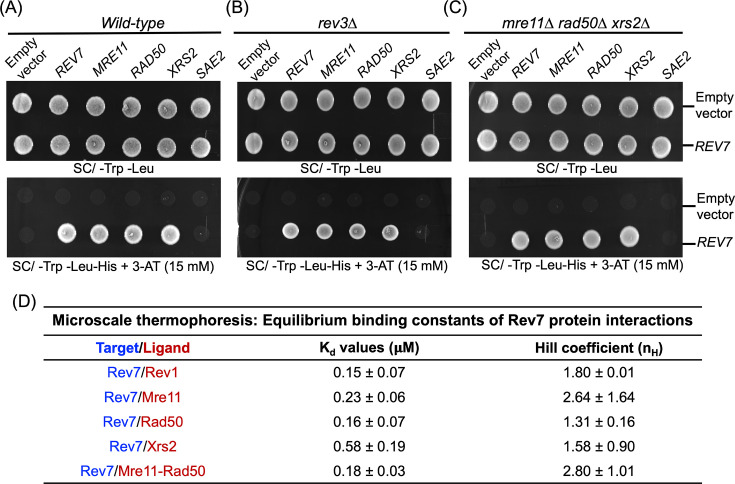
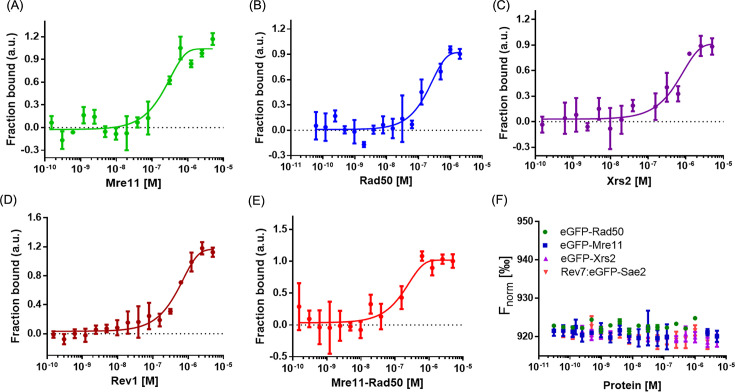
Figure 1. Y2H screens suggest interaction between ScRev7 and the MRX subunits.
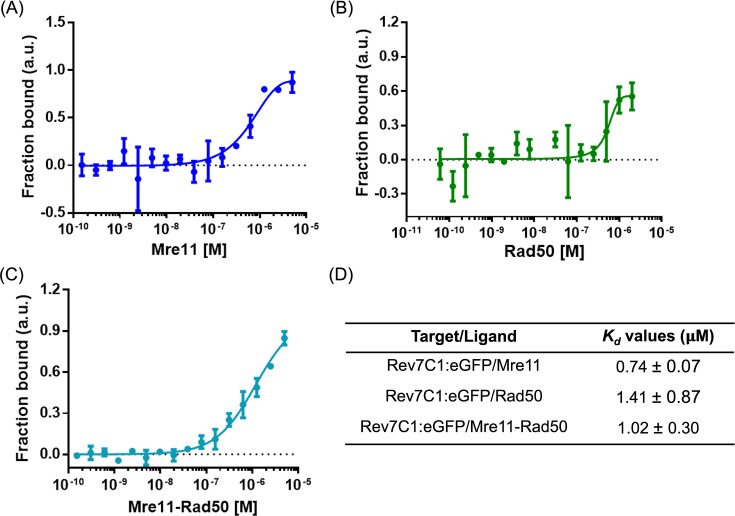
The Y2H assay was performed in (A) wild-type, (B) rev3Δ, and (C) mre11Δ rad50Δ xrs2Δ mutant strains in PJ69-4A background. These strains were co-transformed with pairwise combinations of empty vector, bait (pGBKT7-REV7) and prey (pGADT7/MRE11, RAD50, XRS2, REV7, or SAE2) plasmids. Equal number of mid-log phase cells was spotted onto the SC/-Trp-Leu agar plates (upper panels) or SC/-Trp -Leu-His agar plates containing 3-aminotriazole (3-AT) (bottom panels). Cells were imaged after 48 hr of growth at 30°C. The images shown in panels (A–C) are representative of three independent experiments. (D) Quantitative parameters for interaction between ScRev7 and Rev1, Mre11, Rad50, Xrs2, or Mre11–Rad50 proteins.