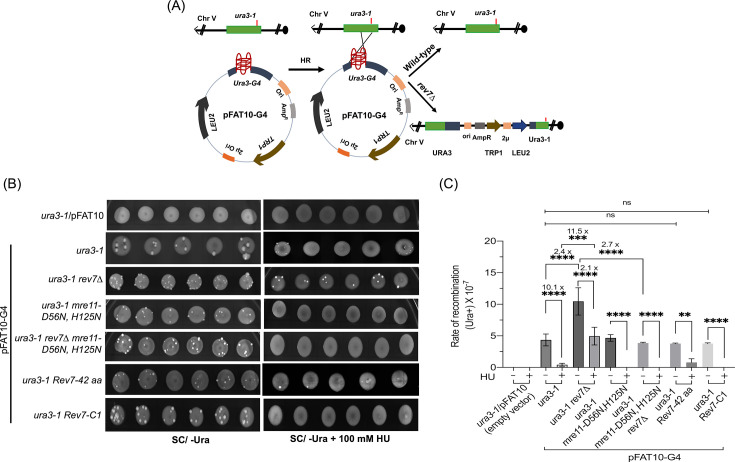
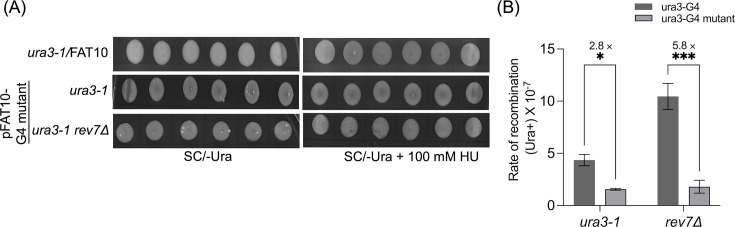
Figure 9. Deletion of REV7 increases the frequency of mitotic homologous recombination (HR).
(A) Schematic representation of plasmid-chromosome recombination assay. The ura3-1 and ura3-G4 alleles are located on chromosome V and plasmid pFAT10-G4, respectively. Recombination between a plasmid borne ura3-G4 allele and the chromosomal borne ura3-1 allele would result in Ura+ prototrophs. (B) Representative images of Ura3+ papillae on SC/-Ura agar plates in the absence or presence of 100 mM HU. (C) Quantification of the rate of HR frequency in different strains. Data are presented as mean ± SD from three different experiments. ns, not significant, **p < 0.01, ***p < 0.001, ****p < 0.0001 versus control, as assessed using one-way ANOVA and Tukey’s post hoc test.