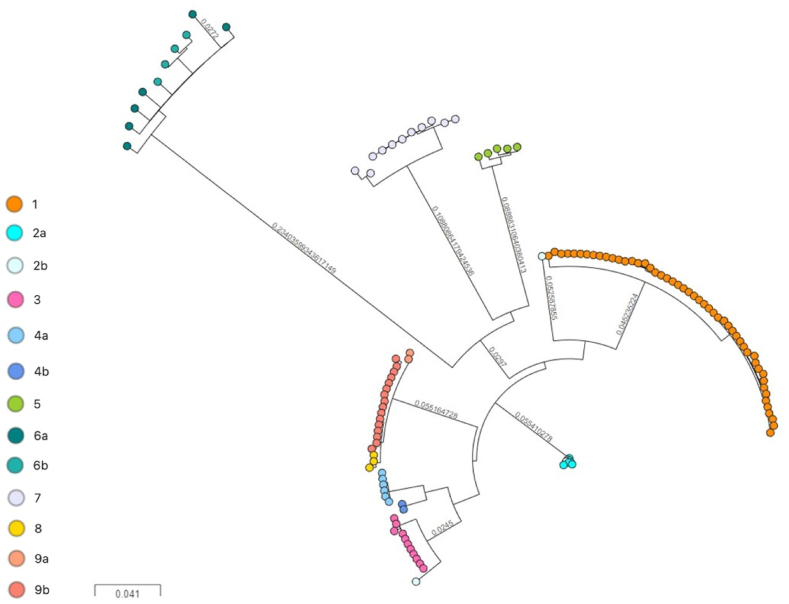
Fig. 2.
wgSNP-based phylogeny of 199 M. tuberculosis complex isolates using a threshold of 5 SNPs to define clusters [excluding Cluster 10 (n = 32) published in Ref. [31]; and 9 other isolates from clusters 1 (n = 5), 2a (n = 2), 2b (n = 1) and 4b (n = 1)]. The final SNP tree was generated by RAxML and visualised with MicroReact. Regions annotated as repetitive elements, InDels, multiple consecutive SNPs in a 12-bp window, and 92 genes implicated in antibiotic resistance were excluded for the phylogenetic reconstruction. Isolates are coloured according to their MIRU-VNTR cluster.