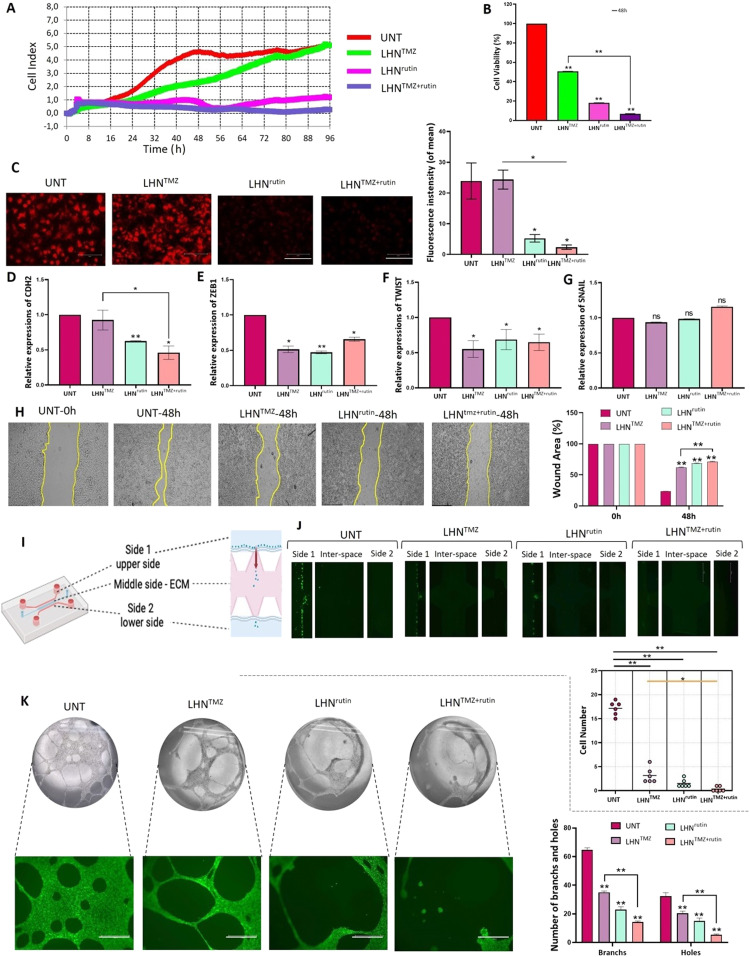
Fig. 2.
The effects of LHNTMZ, LHNrutin and LHNTMZ+rutin on T98 G cell growth and aggressiveness. T98 G cells were treated with 0.5 cm2 of LHNs encapsulated with IC50 doses of TMZ (1,000 µM), rutin (142.2 µM) and their combination. (A) The effects of LHNTMZ, LHNrutin, and LHNTMZ+rutin treatments on real-time cell proliferation of T98 G cells over 96 h (B) The T98 G cell viability percentage after 48-h treatment with LHNTMZ, LHNrutin and LHNTMZ+rutin (**P < 0.001). (C) The effect of LHNTMZ, LHNrutin and LHNTMZ+rutin on Δψm in T98 G cells. TMRE accumulation of cells (red color) is a sign of high membrane potential. Depolarized mitochondria result in reduced TMRE accumulation. (D) RNA expression of EMT-signaling gene, CDH2. (E-G) RNA expression of EMT regulator transcription factors ZEB1, TWIST and SNAIL. For D-G: Ct values were normalized to the Ct of GAPDH. (*P < 0.05, **P < 0.0001). (H) The representative images of the findings on the scratch wound healing assay. 90 % confluent cell monolayer scratched and recorded as the time 0. The change in the size of the wounded area was recorded after 48-h of treatment with LHNTMZ, LHNrutin or LHNTMZ+rutin. Scale bar: 595.2 µm. (I) The schema of the microfluidic invasion model on an IC—Chip. (J) The movement of GFP-expressing T98 G cells from side 1 (upper side) to side 2 (lower side) and their invasion to the EMC at the interspace was observed by fluorescent microscopy over 48 h 10 % FBS was used as the chemoattractant. Scale bar: 750 µm. *P < 0.05, **P < 0.0001. (K) The effect of the TMZ, rutin and TMZ+rutin release of LHN on tube forming of HUVEC cells. Calcein AM confirmed the viability of HUVEC cells of the capillary structure during decomposition by green color. Scale bar: 750 µm. **P < 0.0001 CDH-2: N-cadherin, ZEB-1: zinc finger E-box binding homeobox 1, TWIST-2: Twist-related protein, SNAIL: zinc finger protein SNAI1. The number of biological replicates was A-H:3, J:6 and K:5. All values were expressed as the mean ± SD (standard deviation) of the biological replicates. All P values were presented for comparison with the UNT samples.