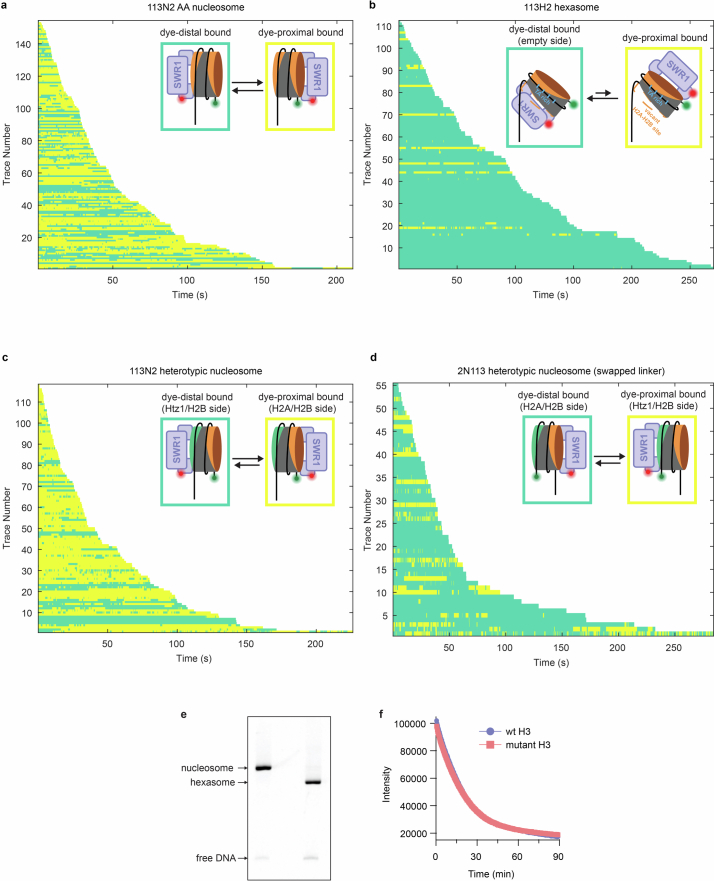
Extended Data Fig. 5. Rastergrams summarising the nucleosome flipping data for different nucleosomes/hexasome, and preparation of yeast hexasomes.
a-d, Each horizontal line represents a smFRET trajectory, ordered by photobleaching/dissociation time. Color indicates whether SWR1 is bound in the dye-distal (green) or dye-proximal (yellow) orientations. Thresholding (at 0.25 FRET) of the idealized FRET trajectories was used to determine the two states. Data is shown for: a, Canonical H2A–H2B 113N2 nucleosomes. b, Hexasomes 113H2 containing only one H2A–H2B dimer. c, Heterotypic nucleosomes 113N2 containing both H2A–H2B and Htz1–H2B histones. d, Swapped linker heterotypic nucleosomes 2N113 containing both H2A–H2B and Htz1–H2B histones. e, Native PAGE comparing a nucleosome and hexasome sample. Representative gel of two independent preparations. f, H3MPQ mutations required for formation of S. cerevisiae hexasomes and heterotypic nucleosomes (see Methods) have no effect on SWR1 exchange activity as measured by bulk FRET decrease.