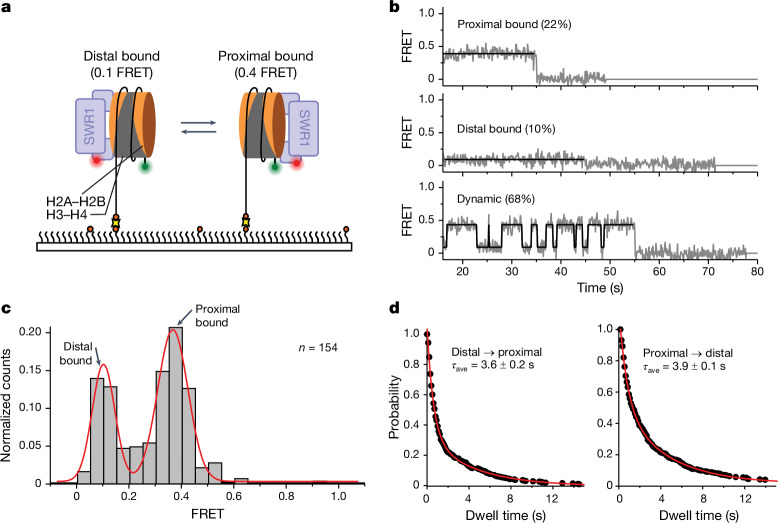
Fig. 3. SWR1 flips between each face of a nucleosome.
a, Schematic of the assay. Nucleosomes (113N2.Cy3) labelled with Cy3 on the short 2-bp overhang are surface immobilized on a PEGylated microscope slide. SWR1, labelled with Atto647N on the N terminus of the Arp6 subunit (SWR1(647N)) is flowed in and allowed to bind to the nucleosome. Interactions between the nucleosome and SWR1(647N) are monitored via smFRET between the donor (green circle) and acceptor (red circle). b, Examples of typical smFRET (grey) and idealized (black) traces. Some molecules display a static FRET of either 0.4 or 0.1, whereas other molecules dynamically flip between these two FRET states. c, Idealized FRET histogram shows two major populations of SWR1(647N) bound to a nucleosome: a low-FRET (0.1) population corresponding to SWR1(647N) bound to the dye-distal side of the nucleosome, and a mid-FRET (approximately 0.4) population corresponding to SWR1(647N) bound to the dye-proximal side of the nucleosome. d, Dwell time plots for the distal-to-proximal (left) and proximal-to-distal (right) transition. The average dwell times (τave) for SWR1 bound in the distal and proximal orientations are approximately equal. Reported errors are the error of the fit.