Fig. 1. Sequences partially similar to ZntR recognition motifs are located near Zur-binding sites in 10.1038/s41467-024-55017-z Zur-regulon promoters.
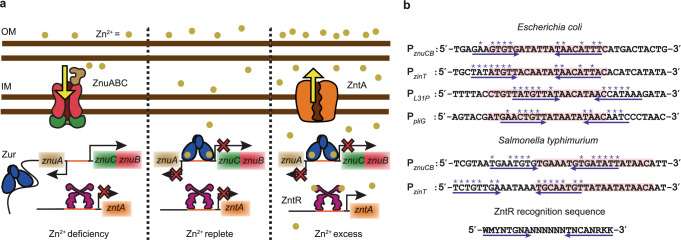
a Present functional paradigm of Zur and ZntR in bacteria. Left: Under Zn2+ deficiency, Zur, without metallation at its regulatory sites, is a non-repressor that binds to non-consensus DNA sites but not to its recognition sequence (i.e., Zur box) in the promoter regions of its regulons (e.g., the divergent znuABC operon); here Zn2+ uptake genes are derepressed, while the non-metallated ZntR binds to its recognition sequence in the promoter regions of its regulons (e.g., zntA), repressing Zn2+ efflux. Center: Under Zn2+ replete conditions, Zur starts to be fully metallated (ZurZn) and binds to Zur box, repressing Zn2+ uptake, while ZntR is still predominantly in its apo form, repressing Zn2+ efflux. Right: Under Zn2+ excess, fully metallated ZurZn keeps repressing Zn2+ uptake, while the metallated ZntRZn at its cognate promoters distorts the DNA to result in activation of Zn2+ efflux genes. Both Zur and ZntR also have a freely diffusing population in the cell (not shown). IM inner membrane, OM outer membrane. b Promoter region sequences of Zur regulons in two different bacteria. Pink shades: Zur boxes. Double blue arrows: possible dyad symmetric sequences recognized by ZntR, whose consensus recognition sequence is shown at the bottom. Asterisk (*) denotes matches with the consensus sequence. Analysis of other species in Supplementary Notes 1.