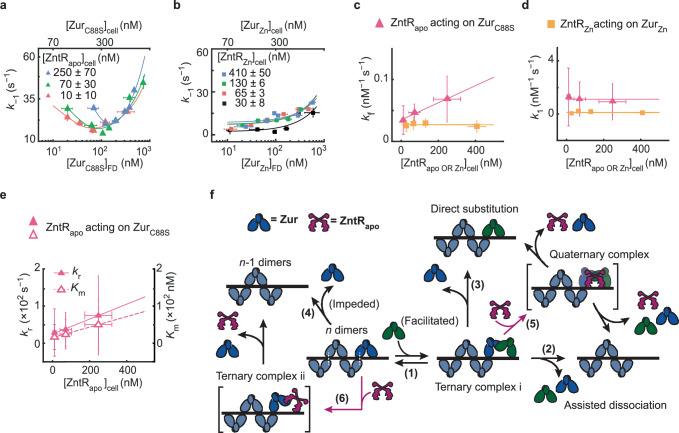
Fig. 5. ZntR effects on Zur-DNA interactions.
a Dependence of the apparent unbinding rate constant k−1 of on its own concentration and at different [] in the cell (orange: n = 514 cells; green: n = 642 cells; blue: n = 532 cells). Lines: fits with Eq. (1). b Same as (a), but for k−1 of and at different [] in the cell (black: n = 611 cells; orange: n = 287 cells; green: n = 494 cells; blue: n = 623 cells). Lines: fits with Eq. (1) including 1st and 3rd terms only. c The facilitated unbinding rate constant kf of vs. cellular [] (magenta triangle) and of vs. the cellular [] (yellow square) (n = 1978 cells). Lines: linear (magenta) and horizontal line (yellow) fits. d Same as (c) but for the binding rate constant k1. Lines: horizontal line fits (n = 2039 cells). e The impeded unbinding rate constant kr (solid triangle) and the effective oligomer dissociation constant Km (open triangle) of vs. cellular [] (n = 2039 cells). Lines: linear fits. Error bars in (a–e) are SEM. f Mechanistic model for ZntRapo-dependent Zur unbinding kinetics. Starting with oligomerized Zur (dark and light blue) at a tight-binding site on DNA, the unbinding of an incumbent Zur protein (dark blue) can be facilitated by a freely diffusing Zur (dark green) through the formation of a ternary complex i (step 1), leading to assisted dissociation (step 2) or direct substitution (step 3); this facilitated unbinding of Zur can be enhanced by ZntRapo through the formation of a heteromeric quaternary complex (step 5). The oligomer-induced impedance of Zur unbinding (step 4) can be weakened by ZntRapo through the formation of a heteromeric ternary complex ii (step 6), leading to faster Zur unbinding as well. White dashed lines on the ‘n dimers’ denote salt bridge interactions between Zur dimers. Source data are provided as a Source Data file and also available in Supplementary Table 7.