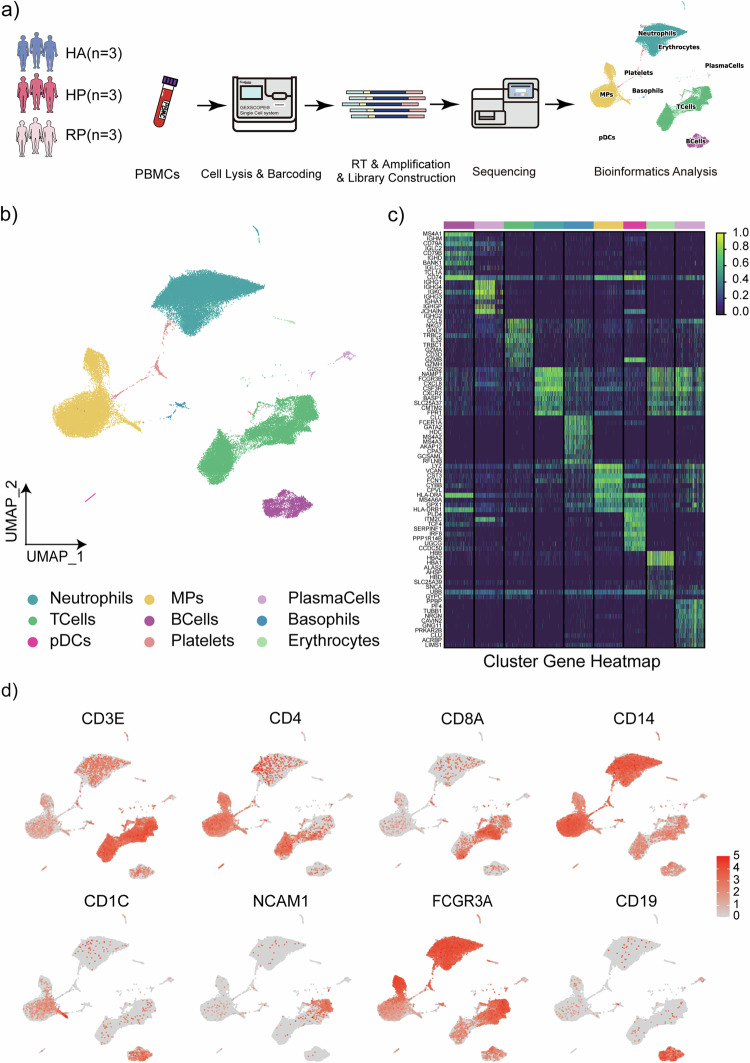
Fig. 1. Study design and analysis of single immune cell profiling in the onset of and recovery from herpes zoster.
a Schematics of the experimental design for single-cell RNA (sc-RNA) sequencing. Peripheral blood mononuclear cells (PBMCs) were processed via sc-RNA sequencing using the 10x-Based Genomics platform after collected from herpes zoster patients (HP), herpes zoster patients after recovery (RP) and healthy controls (HA). b UMAP plot showing eight subpopulations of PBMCs including neutrophils, MPs, plasma cells, T cells, B cells, basophils, CDCs, platelets and erythrocytes identified using integrated and classification analysis. c The heatmaps show differentially expressed genes (DEGs) upregulated in eight subpopulations of PBMCs. d UMAP projection of canonical markers, including CD3E, CD4, CD8A, and NCAM1 for NK and T cells; CD14, CD1C, and FCGR3A for myeloid cells; and CD19 for B cells as indicated in the legend.