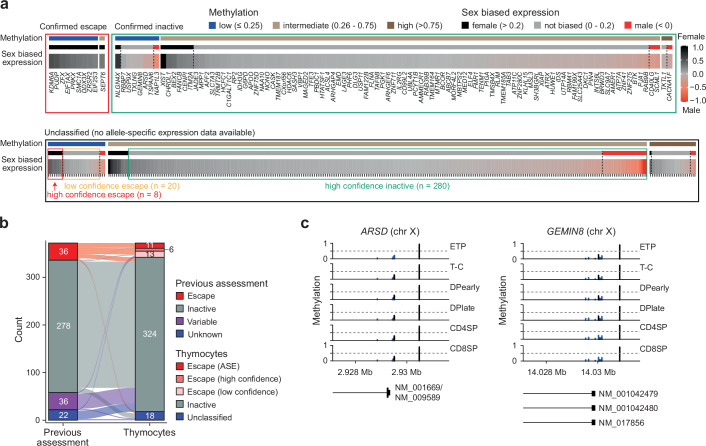
Fig. 7. X-inactivation during T cell development.
a X-inactivation escape status as inactive (green box) or escape (red and orange box), DNA methylation (low, intermediate or high) and sex-biased expression as fold change (FC) (log2) in females over males of all expressed chromosome X genes in thymocytes. Confirmed escape status is inferred from F3 allele-specific expression analysis (inactive genes = mean allele-specific expression across thymocyte development (ASE) >0.4, escape genes = ASE < 0.4). DNA methylation for each gene as mean methylation of −500 TSS probes categorized into low (methylation ≤0.25), intermediate (0.26–0.75) and high ( >0.75) methylation. High confidence escape genes: low promoter methylation and sex-biased expression > 0.2; low confidence escape genes: low promoter methylation and sex biased expression 0-0.2; high confidence inactive genes: intermediate or high methylation and sex-biased expression < 0.2. Genes that fall outside of these thresholds are considered unclassified. See “methods” for details. b Escape status comparison between previous escape assessment10 and our new classification in thymocytes. Calls as escape genes were either based on allele-specific expression (ASE) or as high/low confidence according to panel a. c, DNA methylation of probes around transcription start sites of X-chromosome genes, ARSD and GEMIN8. ETP, early T cell progenitors; T-C, T cell committed thymocytes; DPearly, early double positive thymocytes; DPlate, late double positive thymocytes; CD4SP, CD4 single positive thymocytes; CD8SP, CD8 single positive thymocytes. Expression data from seven biological replicates (4 female, 3 male) and DNA methylation data from three female biological replicates. Source data are provided as a Source Data file.