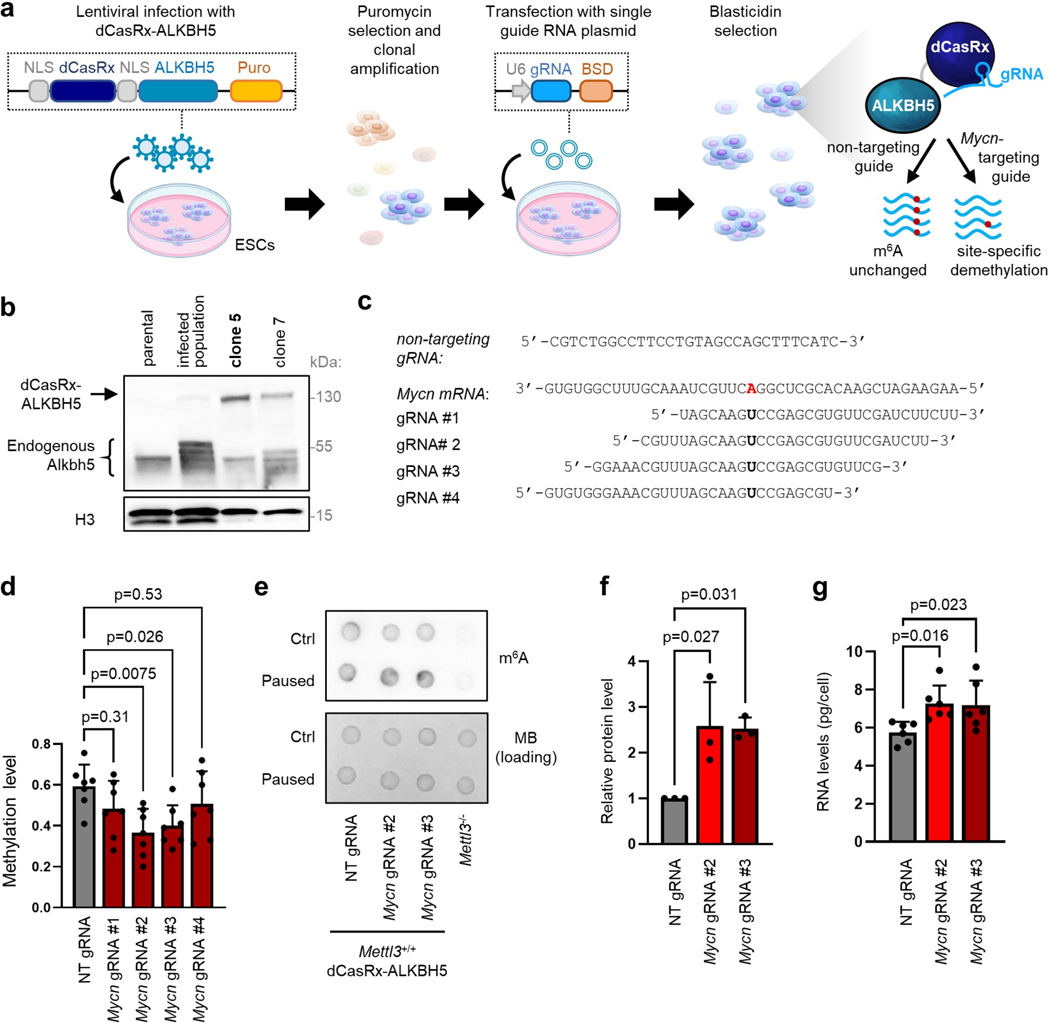
Extended Data Figure 9. Targeted m6A demethylation controls expression of Mycn in paused ESCs.
a. Model of lentiviral dCasRx epitranscriptomic editor with the m6A demethylase ALKBH5 and single guide RNA. b. Validation of the expression of the dCasRx-ALKBH5 fusion by western blot in parental (non-infected) ESCs, infected ESCs, and 2 infected clones. Clone 5 was used for all experiments (representative of 2 biological replicates). c. Guide RNAs (gRNAs) transfected for non-targeting control and Mycn-targeting conditions. d. Changes in m6A using the dCasRx-ALKBH5 editor in paused ESCs. Guides #2 and #3 significantly reduce m6A in Mycn transcripts, as measured by m6A-qPCR, and were selected for all subsequent experiments. N=7 biological replicates. NT: non-targeting. e. Dot blot showing that the global increase of m6A in paused ESCs is not affected by the dCasRx-ALKBH5 editor, with Mettl3−/− ESCs as negative control. Representative of 3 biological replicates. MB: methylene blue. f. Quantification of N-Myc protein levels, showing increased expression with the dCasRx-ALKBH5 editor targeting Mycn in paused ESCs, with representative blot shown in Fig. 6e. N=3 biological replicates. g. Demethylation of Mycn increases the total RNA levels per cell in paused ESCs (n=6 biological replicates).
Data are mean ± SD (d, f-g) and P-values (as indicated on figure) by one-way ANOVA with Dunnett’s multiple comparison tests (d, f-g).