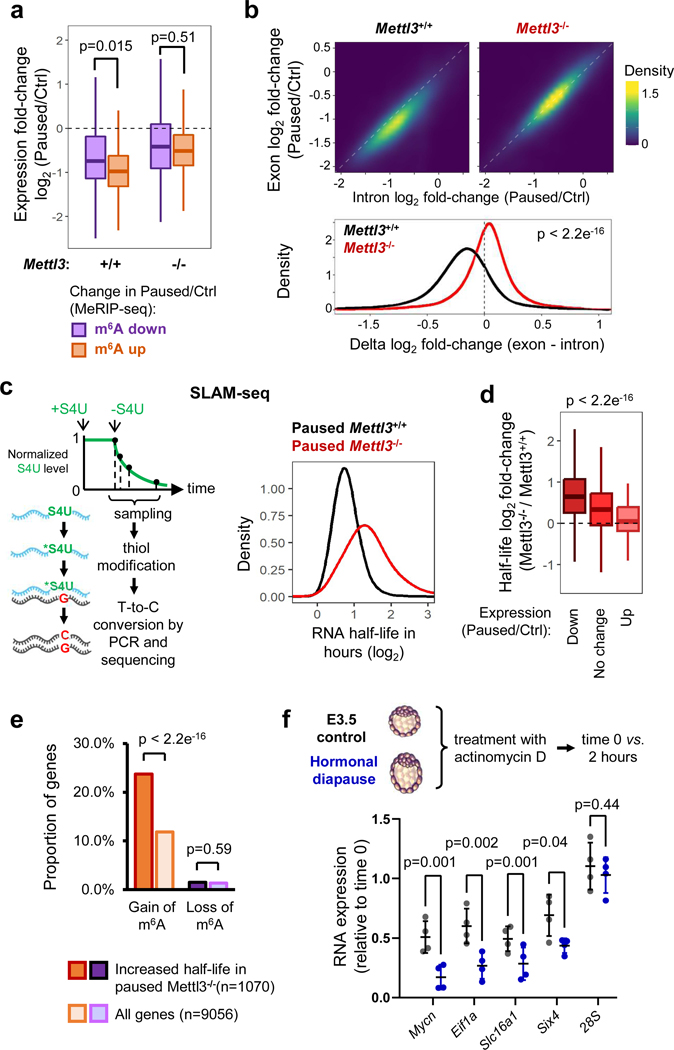
Figure 4: Mettl3 promotes RNA destabilization during pausing.
a. RNAs with increased m6A in pausing (as defined in Fig. 3e) are significantly more downregulated than RNAs with decreased m6A. This pattern is Mettl3-dependent, as analyzing the same RNAs in Mettl3−/− ESCs shows no effect. RNA-seq data as shown in Fig. 2f (n=3 biological replicates per group).
b. Differences in expression (log2FC paused/Ctrl) between exonic and intronic RNA-seq data indicate a global decrease in RNA stability in Mettl3+/+ ESCs upon pausing. This effect is absent in Mettl3−/− ESCs. c. Schematic of the measurement of RNA degradation kinetics by SLAM-seq (left). In the paused state, Mettl3−/− ESCs display an overall longer half-life of the transcriptome compared to Mettl3+/+ ESCs (right). Half-lives were measured using 4 time points, with samples collected over 2 independent experiments (see Extended Data Fig. 5d). S4U: 4-thiouridine. d. Changes in RNA expression during pausing in wild-type ESCs (Paused/Ctrl, fold-change > 1.5) are anti-correlated with changes in RNA half-life in paused Mettl3−/− ESCs (as measured in Fig. 4c). e. RNAs with increased m6A in pausing (as defined in Fig. 3e) are enriched among RNAs stabilized in Mettl3−/− ESCs (half-life fold-change > 1.5). f. Increased RNA stability in control E3.5 blastocysts compared to diapaused blastocysts, as measured by treatment with actinomycin D for 2 hours followed by RT-qPCR (n = 4 biological replicates). Ribosomal 28S as negative control for RNA decay. All data are mean ± SD.
P-values (as indicated on figure) by two-tailed Student’s t-tests (a-b, f), one-way ANOVA (d) and two-proportion z-tests (e). Boxes in the box plots define the interquartile range (IQR) split by the median, with whiskers extending to the most extreme values within 1.5 × IQR beyond the box.