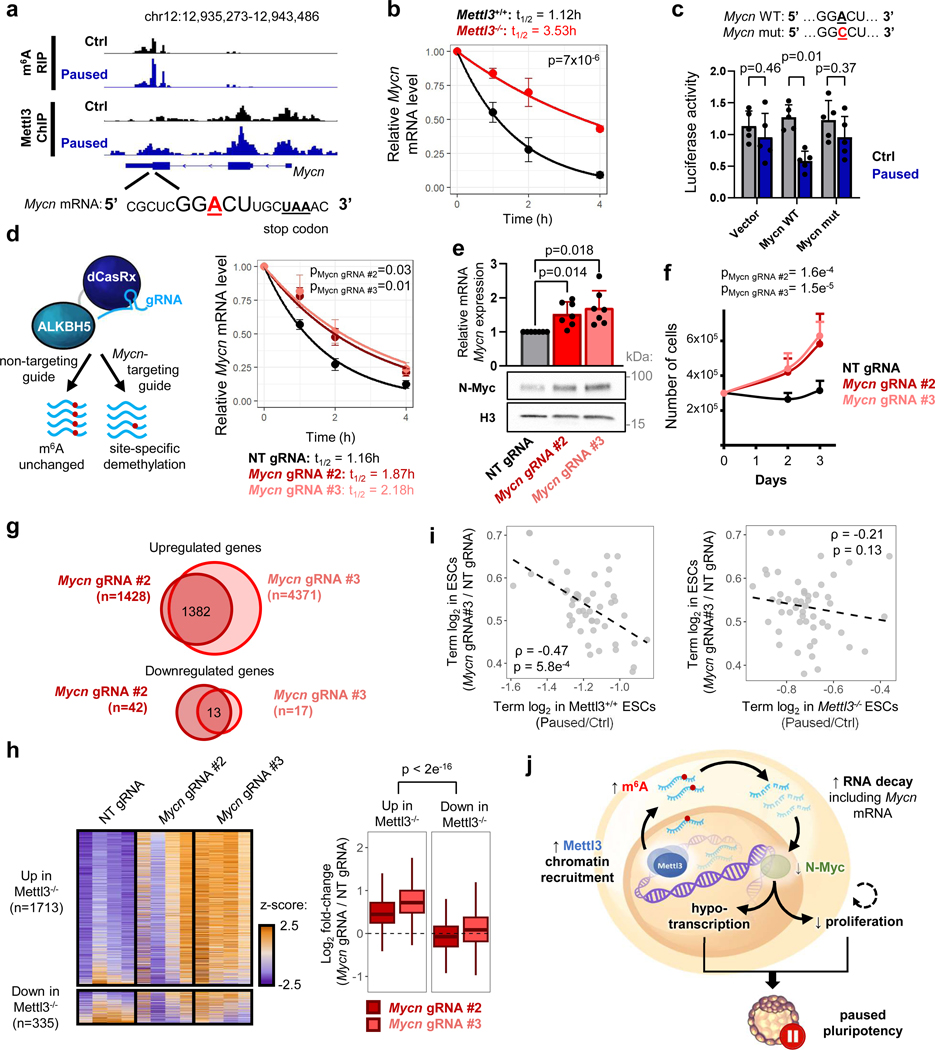
Figure 6: Mettl3 regulates pausing via m6A-mediated destabilization of Mycn mRNA.
a. Gene track view of MeRIP-seq and Mettl3 ChIP-seq for Mycn mRNA. b. Increased Mycn mRNA stability in paused Mettl3−/− ESCs compared to paused Mettl3+/+ ESCs, as measured by an actinomycin D stability assay (n=3 biological replicates). t1/2: half-life. c. Insertion of the identified Mycn m6A site, but not its mutated version, reduces transcript stability in paused ESCs, as measured by a luciferase reporter assay (n=5 biological replicates). d. Site-specific demethylation of Mycn, achieved with a dCasRx conjugated to the m6A demethylaseALKBH5, directed by gRNAs (left), leads to increased Mycn mRNA stability (right, n=3 biological replicates). NT: non-targeting. e-f. Site-specific demethylation of Mycn phenocopies Mettl3−/− in paused ESCs, with increased expression of N-Myc by RT-qPCR (e top, n=7 biological replicates) and western blot (e bottom, representative of 3 biological replicates), and higher proliferation (f, n=5 biological replicates). g. Differential gene expression, as measured by RNA-seq, induced by targeted demethylation of Mycn mRNA in paused ESCs (n=4 biological replicates per condition, fold-change > 1.5 and adjusted P < 0.05). h. Genes upregulated in paused Mettl3−/− ESCs (compared to paused Mettl3−/− ESCs) are also significantly upregulated following the demethylation of Mycn mRNA. Number of genes (n) as indicated. Data as z-score normalized per gene, with all samples displayed (n=4 biological replicates per group, left) and log2 fold-change over NT gRNA control (right). (i) Scatter plots of the median log2 fold-changes for each “hallmark” gene set, showing a significant negative correlation between paused ESCs with demethylated Mycn mRNA and paused Mettl3+/+ (but not Mettl3−/−) ESCs. Spearman correlation coefficient (ρ) is indicated. j. Model for the role of Mettl3-dependent m6A methylation in paused pluripotency. Elevated chromatin recruitment of Mettl3 increases m6A in the transcriptome. Hypermethylation destabilizes many transcripts, including the mRNA encoding the “anti-pausing” factor N-Myc. In absence of Mettl3, upregulated N-Myc enhances transcription and proliferation, disrupting pausing.
All data are mean ± SD (c, e-f) or mean ± SEM (b, d). P-values (as indicated on figure) by linear regression test with interaction (b, d, f), two-tailed paired Student’s ratio t-tests (c), one-way ANOVA with Dunnett’s multiple comparison tests (e), two-way ANOVA (h), two-sided Spearman correlation test (i). Box plots present center lines as medians, with box limits as upper and lower quartiles and whiskers as 1.5×IQR.