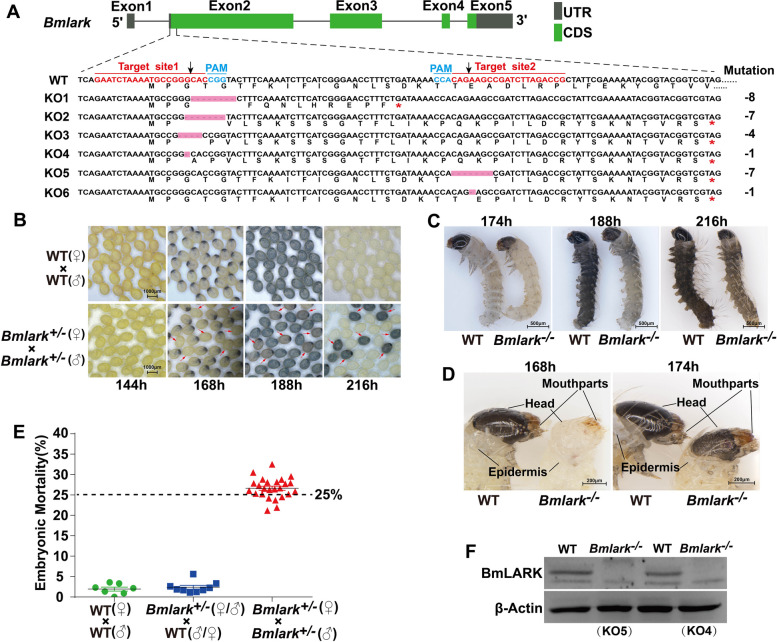
Fig. 1.
CRISPR/Cas9-mediated Bmlark knockout and the phenotypes of Bmlark knockout mutants. A Schematic diagram of the Bmlark gene, target sites, and mutation types. UTR: untranslated regions; CDS: coding sequence; red asterisks represent the stop codon; pink boxes represent missing sequences; sequences in red are the target sites; sequences in cyan are the protospacer adjacent motif (PAM); black arrows indicate the positions at which Cas9 cleaved the target DNA. WT: wild-type sequence; KO1-KO6: mutant sequences. B Developmental difference of Bmlark knockout eggs of the G2 generation 144–216 h post oviposition. Red arrows indicate the Bmlark−/− eggs. WT: wild-type; time (h) is after oviposition. C Phenotypes of the WT and homozygous Bmlark−/− embryos at 174, 188 and 216 h post oviposition. D Detailed differences in the head and epidermis between WT and Bmlark−/− embryos 168 and 174 h post oviposition. E Embryo mortality of offspring of different genotype crossings. The bars show the means (in black) and standard error (in green, blue, or red) of each group. The results are presented as mean ± SE of multiple biological replicates (Supplementary Table S3). The results of the χ2-test showed that the observed value was not significantly different from the expected value (p > 0.05, Supplementary Table S3), indicating that the mortality rate was consistent with the assumed probability of 25%. F Western blot analysis of BmLARK protein in the WT and Bmlark−/− mutant (KO4 and KO5) embryos at 168 h post oviposition