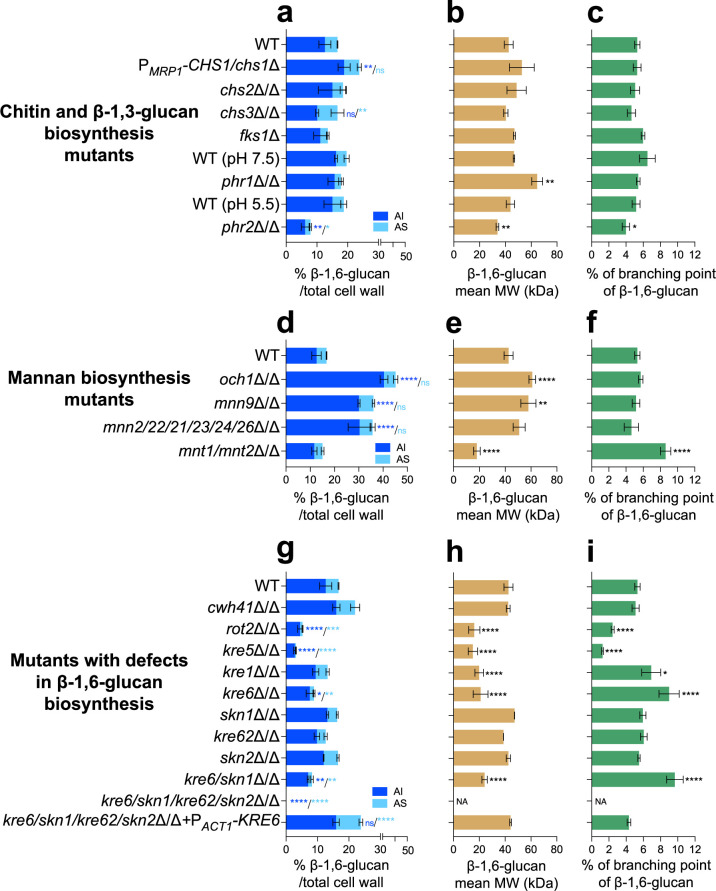
Figure 3. Comparative analysis of β-1,6-glucan content and structure produced by cell wall mutants.
(a, d, g) Percentages of cell wall β-1,6-glucans (alkali-insoluble [AI] and alkali-soluble [AS] fractions) on total cell wall. (b, e, h) β-1,6-Glucans mean molecular weight (MW). (c, f, i) Branching rate of β-1,6-glucans. Cells were grown in liquid synthetic dextrose (SD) medium at 30°C. Means and standard deviations from three independent replicate experiments are shown. All data were compared to the control conditions and were analyzed by one-way ANOVA with Dunnett’s multiple comparisons test: *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, nonsignificant; NA, nonapplicable.
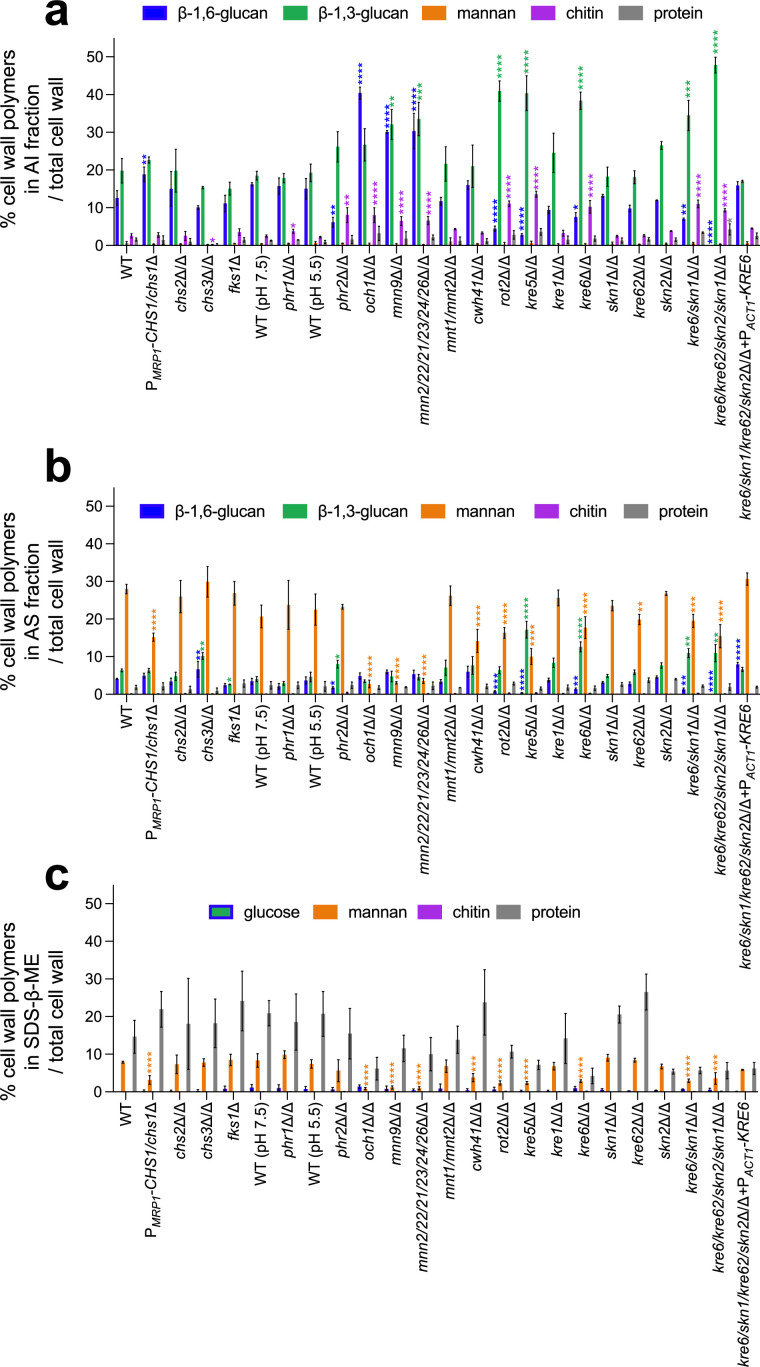
Figure 3—figure supplement 1. Cell wall composition of C. albicans mutants.
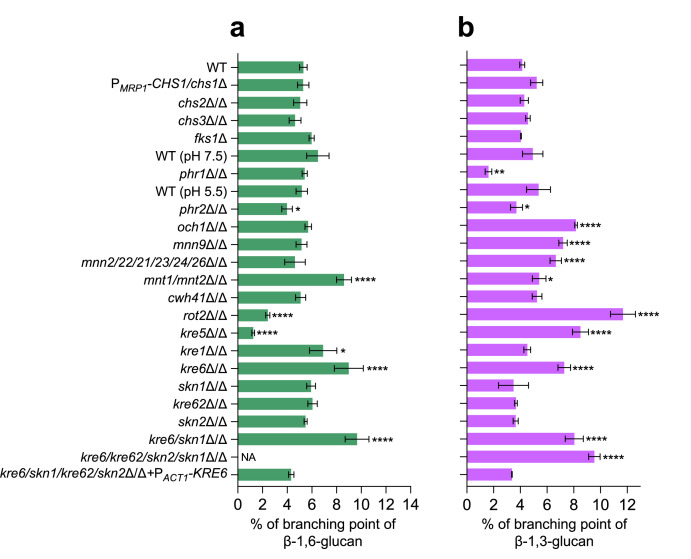
Figure 3—figure supplement 2. Branching rates of β-1,6-glucans and β-1,3-glucans produced by different cell wall mutants of C. albicans.
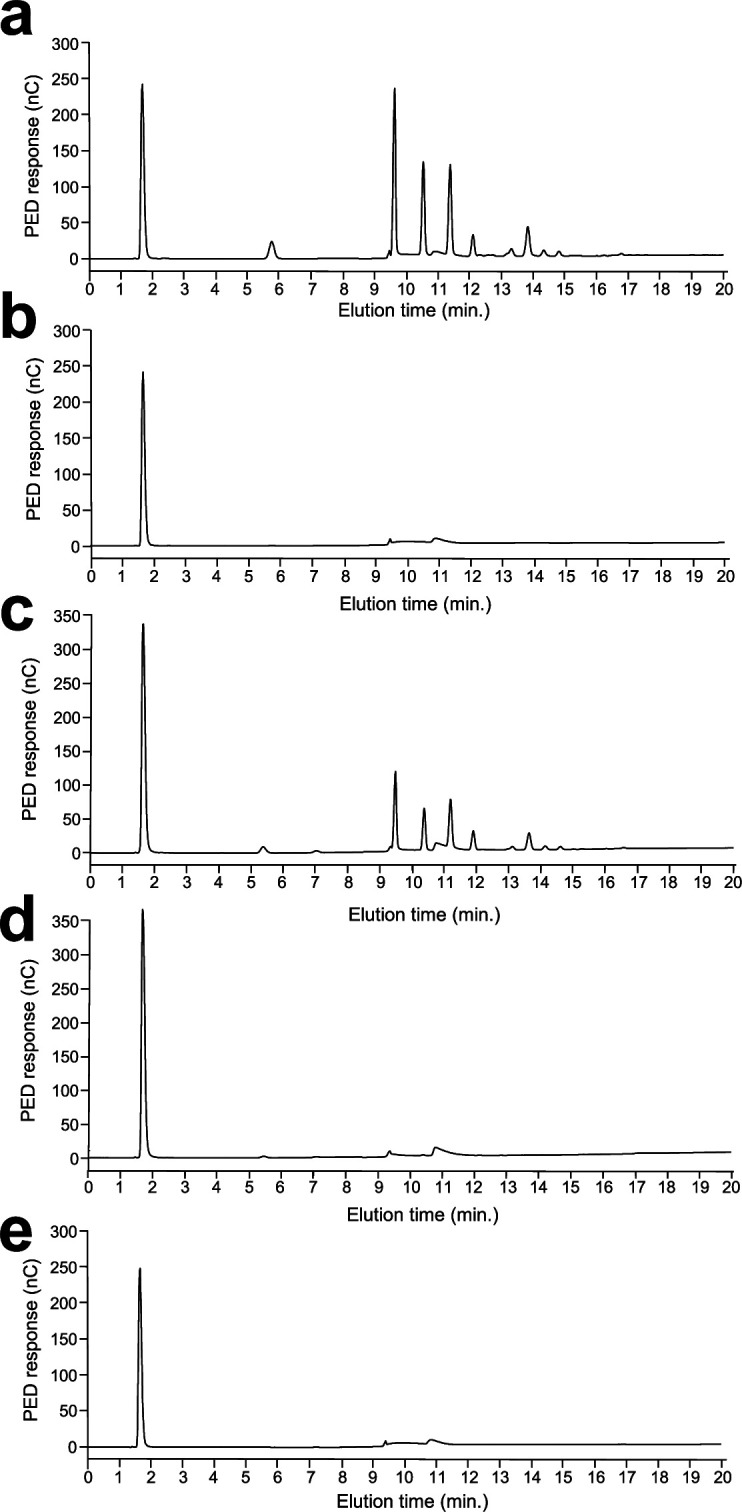
Figure 3—figure supplement 3. Absence of β-1,6-glucans in the cell wall of the quadruple kre6/kre62/skn2/skn1Δ/Δ mutant.