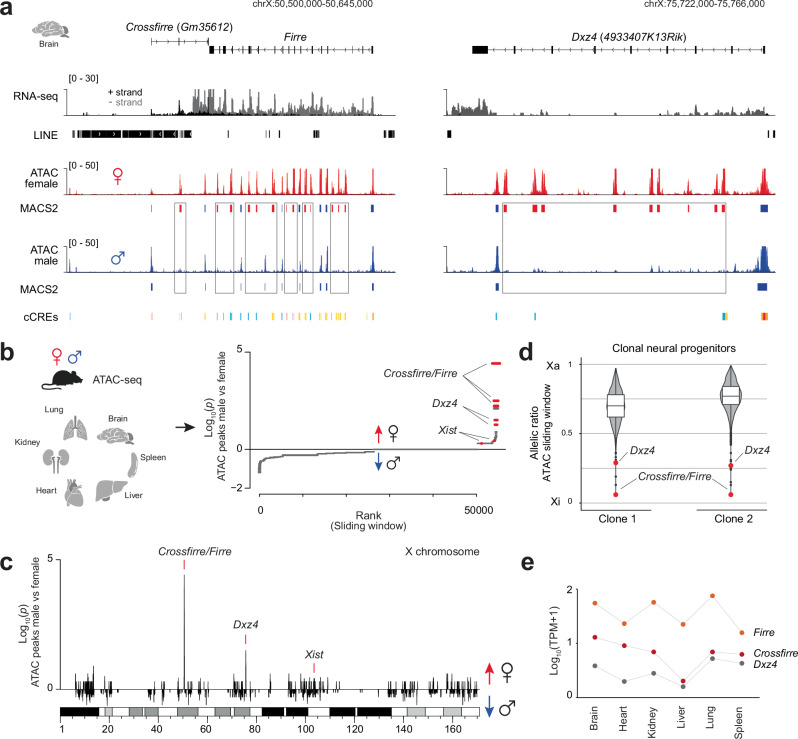
Fig. 1. Crossfirre, Firre, and Dxz4 loci are the topmost female-specific loci by chromatin accessibility with unique allele-specific characteristics.
a Genome browser tracks of the mouse brain showing strand-specific RNA-seq15 and ATAC-seq23 for females (red) and males (blue) covering the Crossfirre (Gm35612), Firre, and Dxz4 (4933407K13Rik) locus. The gene body of Crossfirre is embedded in a 50 Kb LINE cluster. Boxes highlight female-specific ATAC-seq peaks identified through MACS260. ENCODE Candidate Cis-Regulator Elements (cCRES) are shown for each locus (Promoter: red, Proximal enhancer: orange, Distal enhancer: yellow, DNase/H3K4me3: pink, CTCF: blue). b Overview of six female and male organs (two biological replicates per sex, n = 24) utilized to map sex-specific chromatin accessible loci from publicly available ATAC-seq data23 (left). A one-sided binomial test was used to identify sex-specific loci genome-wide by comparing male and female ATAC peak counts binned over 100 Kb sliding windows (right, see section “Methods”). A positive value was assigned if more peaks were present in females than in males, while negative values were assigned if the reverse was true. The top female-specific loci Crossfirre, Firre, and Dxz4 are indicated, together with the expected female-specific Xist locus. c Sex-specific analysis of (b) for the entire X chromosome. d Violin plots displaying the allelic ratio of ATAC enrichment over a 50 Kb sliding window for the entire X chromosome of neuronal progenitor cell clones (n = 2)24. The red dot highlights the allelic ratio of the sliding window overlapping the Crossfirre/Firre and Dxz4 locus. Boxes represent the interquartile range around the median, while the whiskers extend to 1.5 times the interquartile range. e RNA-sequencing abundance of Crossfirre (red), Firre (orange), and Dxz4 (gray) across various mouse organs. Shown are the log10-transformed mean TPM values of each gene within the respective tissue with a pseudocount of 1 added.