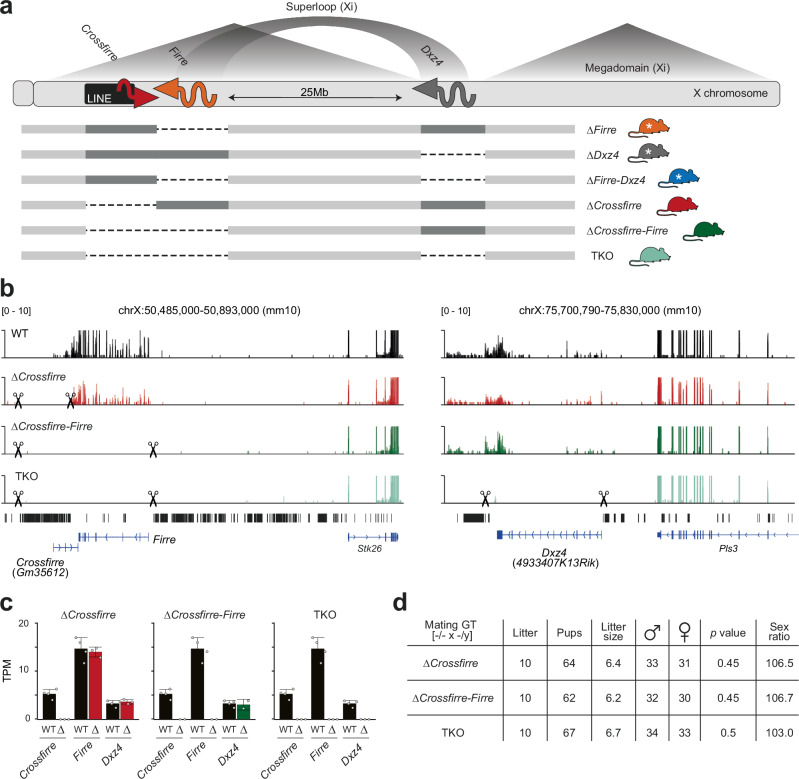
Fig. 2. Mice carrying a Crossfirre single deletion or combined with Firre and Dxz4 are viable and undergo normal development.
a Schematic overview of the X chromosome. The megadomains and the superloop between the Firre and Dxz4 loci are specific to the inactive X chromosome (Xi), whereas full-length transcription of Firre and Dxz4 only occurs on the active X chromosome (Xa). The imprinted lncRNA Crossfirre shows expression from the maternal X chromosome, independent of random XCI15. Dotted lines indicate the deleted loci for ∆Firre (orange), ∆Dxz4 (gray), ∆Firre-Dxz4 (blue), ∆Crossfirre (red), ∆Crossfirre-Firre (green), ∆Crossfirre-Firre-Dxz4 (TKO, turquoise). Colors are used in figures throughout the manuscript to highlight the genotype of origin. White stars refer to previously generated mouse strains14. b Genome browser RNA-seq tracks showing the Crossfirre/Firre (left) and Dxz4 (right) locus for the adult spleen of wildtype (black), and ∆Crossfirre, ∆Crossfirre-Firre, TKO mutants. Scissors indicate the start and end of the CRISPR-Cas9 deletion (see section “Methods”). c Mean expression values of Crossfirre, Firre, and Dxz4 in the adult spleens of Crossfirre mutant strains (WT n = 4, ∆Crossfirre n = 3, ∆Crossfirre-Firre n = 2, TKO n = 3). Error bars indicate the standard deviation. d Sex distribution of homozygous ∆Crossfirre, ∆Crossfirre-Firre, and TKO breeding. The p values are obtained from a one-sided binomial test.