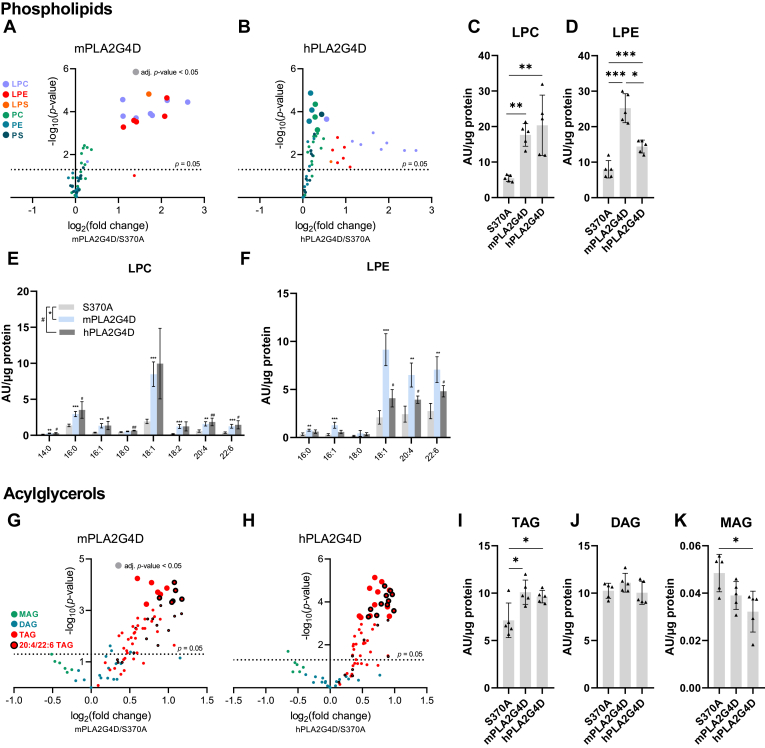
Fig. 5.
PLA2G4D overexpression alters cellular acylglycerol and phospholipid profiles. (A, B) Volcano plots showing changes in glycerophospholipids in m- and hPLA2G4D-expressing COS7 cells compared to control cells expressing the inactive mS370A mutant. The x-axis shows the log2 (fold change) of the indicated phospholipid subspecies. The y-axis shows the -log10(P value). Lipids were analyzed by HPLC-MS 48 h posttransfection. (C, D) Total LPC and LPE levels calculated based on the sum of signals from all detected subspecies. (E, F) Lipid subspecies of LPC and LPE in COS7 cells expressing m-, hPLA2G4D, or the mS370A mutant. (G, H) Volcano plots showing changes of MAG, DAG, and TAG subspecies in m- and hPLA2G4D-expressing cells compared to control cells expressing the mS370A mutant. (I–K) Total MAG, DAG, and TAG levels calculated based on the sum of signals from all detected subspecies. Data are shown as mean ± SD (n = 5) and are representative of three independent experiments. Statistical comparison in (A, B, E, F, G, H) was performed with multiple unpaired two-tailed Student’s t test followed by Bonferroni posthoc analysis. Large-sized spots in volcano plots represent lipids with an adjusted P value < 0.05. Statistical comparisons in (C, D, I, J, K) were performed with one-way ANOVA followed by Bonferroni posthoc analysis. Statistically significant differences are shown as: ∗, #P < 0.05; ∗∗, ###P < 0.01; and ∗∗∗, ###P < 0.001. DAG, diacylglycerol; LPC, lyso-phosphatidylcholine; LPE, lyso-phosphatidylethanolamine; MAG, monoacylglycerol; PLA2G4, phospholipase A2 group IV; TAG, triacylglycerol.