Fig. 9.
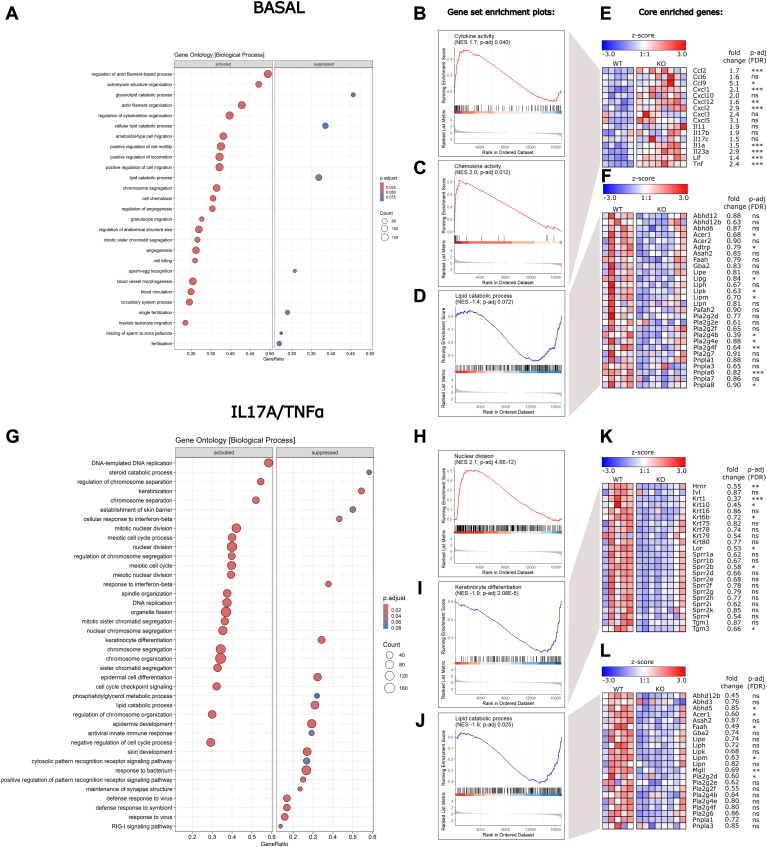
RNA-sequencing of primary WT and Pla2g4d-deficient (KO) keratinocytes. Primary keratinocytes were isolated from two-day-old WT (n = 5) and KO (n = 8) neonatal mice and cultured for four days under low CaCl2 concentrations (0.06 mM). Subsequently, WT and KO cells were stimulated with IL17A and TNFα (20 ng/ml each) for 24 h. (A) “GO biological process” gene set enrichment analysis (GSEA) comparing KO and WT cells in the absence of cytokines (basal). The top 20 activated and 7 suppressed gene sets are shown. The “gene ratio” (x-axis) represents the ratio of core enriched genes (genes before or after the point at which the running enrichment score reaches its maximum or minimum; see B–D and H–J) to the total number of genes in the gene set. The “count” (dot size) reflects the total number of genes in the gene set and the color coding highlights the FDR-adjusted P value of the gene set. (B–D) Gene set enrichment plots of GO molecular function gene sets “Cytokine activity” and “chemokine activity,” as well as the GO biological process gene set “lipid catabolic process” comparing KO and WT cells in the absence of IL17A and TNFα (basal). The plots display the GSEA statistics, including the “normalized enrichment score (NES)” and FDR-adjusted P value. (E, F) Selected core enriched genes from gene sets shown in (B–D). Differential gene expression between KO and WT cells is visualized in a heat map, displaying the z-score (indicates number of SDs a data point is from the mean of the dataset). Additionally, the fold change (KO/WT) and FDR-adjusted P value are shown. (G) “GO biological process” GSEA comparing KO and WT cells in the presence of IL17A and TNFα. The top 20 activated and top 20 suppressed gene sets are shown. (H–J) Gene set enrichment plots of GO biological process gene sets “nuclear division,” “keratinocyte differentiation,” and “lipid catabolic process” comparing KO and WT cells in the presence of IL17A and TNFα. (K, L) Selected core enriched genes from gene sets shown in (I, J). Differential expression between KO and WT cells is visualized in a heat map, displaying the z-score. Additionally, the fold change (KO/WT) and FDR-adjusted P value are shown. Statistically significant differences are shown as: ∗P < 0.1; ∗∗P < 0.01; and ∗∗∗P < 0.001. FDR, false discovery rate; PLA2G4, phospholipase A2 group IV.