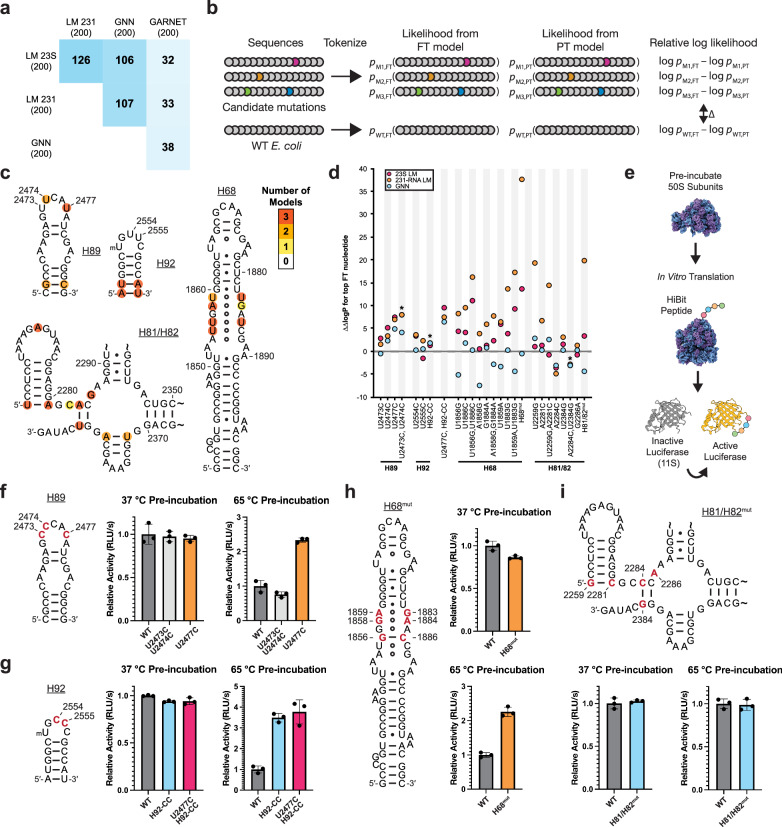
Fig. 6. Mutations in 23S rRNA predicted by deep learning models to confer thermostability on E. coli ribosomes.
a Matrix showing the overlap in the 200 positions with highest Jensen-Shannon divergence in finetuned (FT) model-generated versus pretrained (PT) model-generated sequences for the GNN, LM models, and in the hyperthermophilic versus total GARNET 23S rRNA sequences. b Strategy for calculating ∆∆logP values for candidate mutations, using log likelihoods of sequence generation from FT versus PT models, with WT E. coli serving as a normalization control. c Positions within four regions of the E. coli 23S rRNA with JSD values ranked in the top 200. Coloring indicates the number of models which identify each position. d Analysis of the four regions in panel (c) for candidate thermostabilizing mutations. For each position, the most frequent nucleotide in FT-generated sequences (top FT nucleotide) is grafted into the E. coli 23S rRNA sequence and used to calculate ∆∆logP for the 23S LM, 231-RNA LM, and GNN models. Overlapping values are denoted with an asterisk in the graph for clarity. e Schematic for the heat-treatment in vitro translation assay. Purified 50S subunits are incubated at the indicated temperature, cooled to room temperature, and then added to a HiBit in vitro translation assay. The peptide complements an inactive protein fragment to form an active luciferase. f–i Activity of pre-incubated ribosomes in the HiBit in vitro translation assay. Secondary structures of helices H89 (f), H92 (g), H68 (h), and H81/H82 (i) of E. coli 23S rRNA. Positions that were mutated in this study are shown in red. For panels f through i, WT, and mutant 50S subunits all contain an MS2 tag (Methods). Relative activity is calculated as the slope of the initial increase in luminescence during translation and normalized to the WT value at the given temperature. Data and error bars represent the average and standard deviation of three reactions, respectively. Source data for panels (f) through (i) are provided as a Source Data file.