Abstract
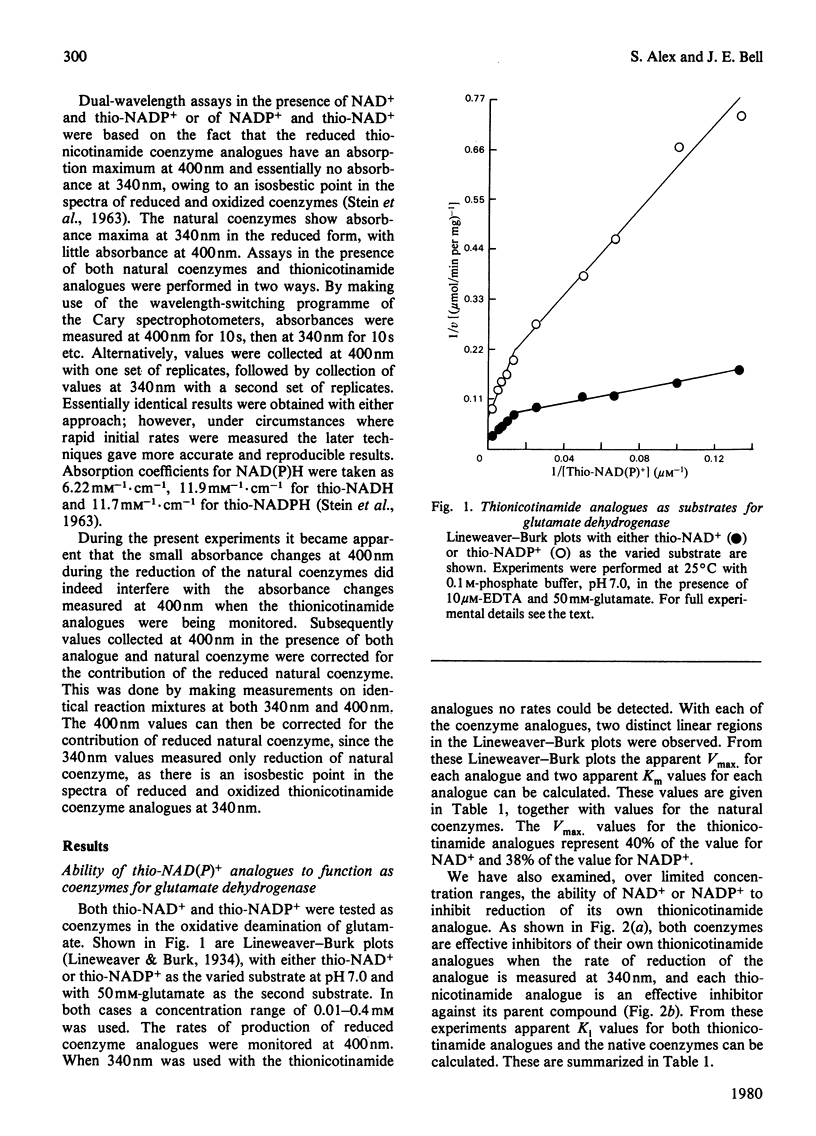
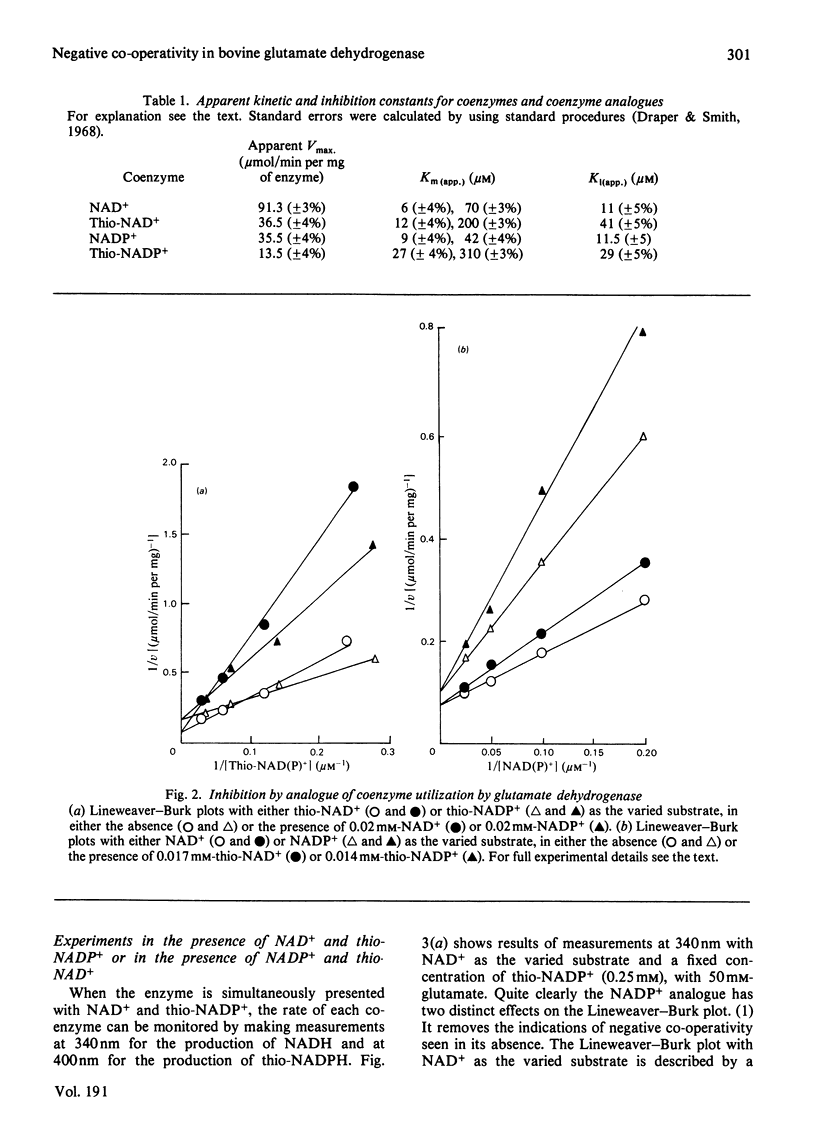
The thionicotinamide analogues of NAD+ and NADP+ were shown to be good alternative coenzymes for bovine glutamate dehydrogenase, with similar affinity and approx. 40% of the maximum velocity obtained with the natural coenzymes. Both thionicotinamide analogues show non-linear Lineweaver-Burk plots, which with the natural coenzymes have been attributed to negative co-operativity. Since the reduced thionicotinamide analogues have an isosbestic point at 340nm and have an absorption maximum at 400nm, it is possible to monitor reduction of natural coenzyme and thionicotinamide analogue simultaneously by dual-wavelength spectroscopy. When glutamate dehydrogenase is presented with NADP+ and thio-NADP+ simultaneously, the enzyme oligomer senses saturation of its coenzyme-binding sites irrespective of the exact nature of the coenzyme and locks the oligomer into its highly saturated form even when low saturation of the monitored coenzyme is present. These experiments substantiate the suggestion that glutamate dehydrogenase shows negative co-operativity in its catalytically active form.
Full text
PDF
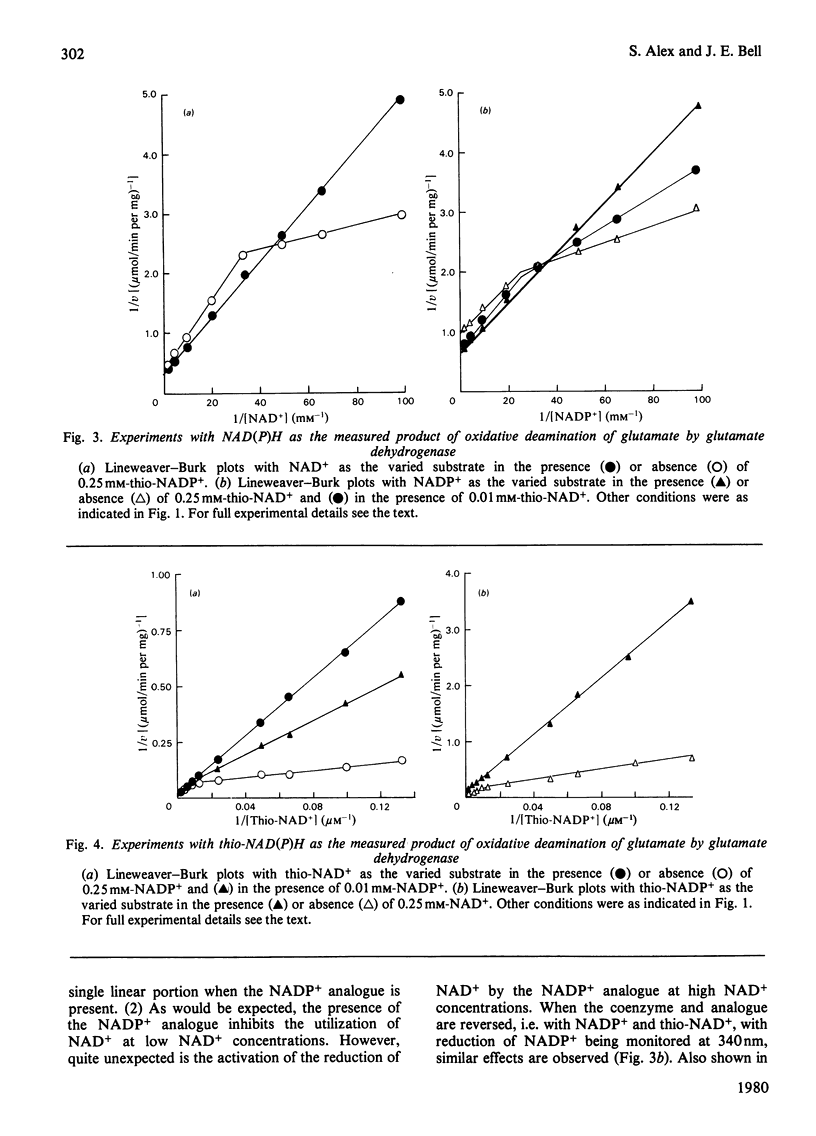


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell J. E., Dalziel K. A conformational transition of the oligomer of glutamate dehydrogenase induced by half-saturation with NAD + or NADP + . Biochim Biophys Acta. 1973 May 5;309(1):237–242. doi: 10.1016/0005-2744(73)90336-7. [DOI] [PubMed] [Google Scholar]
- Cassman M., Schachman H. K. Sedimentation equilibrium studies on glutamic dehydrogenase. Biochemistry. 1971 Mar 16;10(6):1015–1024. doi: 10.1021/bi00782a013. [DOI] [PubMed] [Google Scholar]
- Dalziel K., Egan R. R. The binding of oxidized coenzymes by glutamate dehydrogenase and the effects of glutarate and purine nucleotides. Biochem J. 1972 Feb;126(4):975–984. doi: 10.1042/bj1260975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel Keith, Engel Paul C. Antagonistic homotropic interactions as a possible explanation of coenzyme activation of glutamate dehydrogenase. FEBS Lett. 1968 Oct;1(5):349–352. doi: 10.1016/0014-5793(68)80153-x. [DOI] [PubMed] [Google Scholar]
- Egan R. R., Dalziel K. Active centre equivalent weight of glutamate dehydrogenase from dry weight determinations and spectrophotometric titrations of abortive complexes. Biochim Biophys Acta. 1971 Oct;250(1):47–50. doi: 10.1016/0005-2744(71)90118-5. [DOI] [PubMed] [Google Scholar]
- Engel P. C., Dalziel K. Kinetic studies of glutamate dehydrogenase with glutamate and norvaline as substrates. Coenzyme activation and negative homotropic interactions in allosteric enzymes. Biochem J. 1969 Dec;115(4):621–631. doi: 10.1042/bj1150621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy H. R., Daouk G. H. Simultaneous analysis of NAD- and NADP-linked activities of dual nucleotide-specific dehydrogenases. Application to Leuconostoc mesenteroides glucose-6-phosphate dehydrogenase. J Biol Chem. 1979 Jun 10;254(11):4843–4847. [PubMed] [Google Scholar]
- STEIN A. M., LEE J. K., ANDERSON C. D., ANDERSON B. M. THE THIONICOTINAMIDE ANALOGS OF DPN AND TPN. I. PREPARATION AND ANALYSIS. Biochemistry. 1963 Sep-Oct;2:1015–1017. doi: 10.1021/bi00905a018. [DOI] [PubMed] [Google Scholar]
- Smith E. L., Landon M., Piszkiewicz D., Brattin W. J., Langley T. J., Melamed M. D. Bovine liver glutamate dehydrogenase: tentative amino acid sequence; identification of a reactive lysine; nitration of a specific tyrosine and loss of allosteric inhibition by guanosine triphosphate. Proc Natl Acad Sci U S A. 1970 Oct;67(2):724–730. doi: 10.1073/pnas.67.2.724. [DOI] [PMC free article] [PubMed] [Google Scholar]


