Abstract
Background
Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is one of the most destructive wheat diseases worldwide. Durum wheat (Triticum turgidum L. var. durum Desf.) is a crucial gene donor for improving common wheat.
Results
In this study, we investigated a durum wheat accession, DR88, which exhibits broad and high levels of resistance to powdery mildew. Using bulked segregant RNA-Seq (BSR-Seq), we identified a dominant gene, tentatively designated PmDR88, and localized it to 743–776 Mb interval on chromosome arm 2AL according to the reference genome of durum wheat cv. Svevo. Subsequently, PmDR88 was mapped in a genetic region of 3.9 cM flanked by the markers WGRE77410 and WGRC872 at genetic distances of 1.6 and 2.3 cM, respectively; it also co-segregated with JS717×JS718, the diagnostic marker for the Pm4 locus. Genotyping of a large population comprising 5,174 F2:3 families using JS717×JS718 confirmed that PmDR88 is located at the Pm4 locus on 2AL. Sequence alignment revealed that PmDR88 shares identical amino acid sequences with Pm4d, while qRT-PCR analysis suggested distinct expression patterns for PmDR88 compared with previously reported Pm4 alleles. Two complementary DNA markers, including the dominant co-segregating marker JS717×JS718 and a newly developed closely-linked co-dominant marker WGRE77410, were confirmed to be available for efficiently transferring PmDR88 into the tested wheat backgrounds by marker-assisted selection (MAS) strategy.
Conclusions
PmDR88 was mapped in the Pm4 locus. Despite sharing identical amino acid sequences with Pm4d, PmDR88 exhibits distinct expression patterns. Moreover, DR88 shows broad and high levels of resistance to powdery mildew. Two complementary DNA markers were identified for MAS breeding. The molecular identification of PmDR88 will facilitate transfer of this Pm4 allele into susceptible cultivars for resistance improvement or into resistant cultivars for resistance-enhanced pyramiding breeding.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05884-x.
Keywords: Durum wheat, MAS, Pm4, Powdery mildew
Background
Common wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is an important staple food crop globally, providing approximately 20% of human caloric intake [1]. Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is recognized as one of most devastating diseases affecting wheat production regions worldwide, with potential yield losses ranging from 10 to 15% to up to 62% in severe cases [2, 3].
Although fungicide is often applied in the field management of the disease, utilizing host resistance and breeding resistant cultivars is the preferred method for controlling powdery mildew [4]. To date, more than 60 Pm genes/loci (Pm1-Pm69) have been formally designated from common wheat and its relatives [5]. Up to now, many Pm genes have been cloned from common wheat and its relatives, and most of them encode nucleotide-binding domain leucine-rich repeat (NLR) proteins, including Pm1a (T. aestivum), Pm2 (Aegilops tauschii), Pm3b (T. aestivum), Pm5e (T. aestivum), Pm8 (Secale cereale), Pm12 (Ae. speltoides), Pm17 (S. cereale), Pm21 (Dasypyrum villosum), Pm41 (T. dicoccoides), Pm55 (D. villosum), Pm60 (T. urartu) and Pm69 (T. dicoccoides). Other types of proteins are also identified, such as Pm4b (T. carthlicum) and Pm24 (T. aestivum) encoding kinase-MTC and tandem kinase, respectively, Pm38 (T. carthlicum) and Pm46 (T. aestivum) encoding transporters, Pm36 (T. dicoccoides) encoding membrane associated tandem kinase, Pm13 (Ae. longissima) encoding a mixed lineage kinase domain-like protein and Pm57 (Ae. searsii) encoding a tandem kinase protein [5–10]. However, the direct application of most Pm genes/alleles in wheat breeding programs is hindered by undesirable linkage drag [11, 12]. Even more concerning is the intricate and variable virulence structures exhibited by natural populations of Bgt isolates, which can rapidly overcome host resistance genes [13, 14]. The narrowing genetic diversity of common wheat further complicates breeding efforts [15]. Fortunately, the gene donors are highly diversified, ranging from common wheat to its ancestral species and wild relatives. Although Pm genes derived from wild relatives often confer strong disease resistance, their integration into wheat backgrounds may lead to yield penalties due to the simultaneous introgression of undesirable genes [16]. In contrast, Pm genes derived from ancestral species of common wheat are more likely to balance resistance level and agronomic performance, offering higher breeding potential. Many Pm genes have been identified from the ancestral species of wheat. For example, Pm2, Pm19, Pm34, Pm35 and Pm58 were originally derived from Ae. tauschii (2n = 2x = 14, DD), among which Pm2 have been widely used in more than 100 wheat cultivars in China; Pm60 and Pm25 were derived from T. urartu (2n = 2x = 14, AA) and T. monococcum (2n = 2x = 14, AA), respectively; Pm5a, Pm16, Pm26, Pm30, Pm36, Pm41, Pm42, Pm64 and Pm69 were all derived from T. dicoccoides (2n = 4x = 28, AABB); Pm6, Pm27 and Pm37 were derived from T. timopheevi (2n = 4x = 28, AAGG), among which Pm6 have been widely used in wheat breeding [5]. Therefore, exploring novel Pm genes from these ancestral species is a worthwhile endeavor.
Durum wheat (T. turgidum L. var. durum Desf., simply T. durum, 2n = 4x = 28, AABB) is a key ancestral species of common wheat [17, 18]. Compared with common wheat, it holds more genetic diversity that have not been used in modern wheat breeding, and hence serves as a valuable gene donor for wheat improvement. For powdery mildew resistance, to date, only four Pm genes have been reported in durum wheat: Mld on chromosome 4B [19], Pm3h on 1AS [20], PmDR147 on 2AL [21] and Pm68 on 2BS [22]. Given the untapped potential of durum wheat, we investigated a durum wheat accession, DR88, which exhibited high levels of resistance to powdery mildew during whole growth period. This study aims to (1) determine the inheritance pattern of powdery mildew resistance in DR88, (2) identify the target Pm gene(s) responsible for this race-specific resistance, (3) compare the identified gene(s) with previously reported ones, and (4) screen markers suitable for marker-assisted selection (MAS).
Results
Inheritance of resistance to powdery mildew in DR88
Field trials demonstrated that DR88 exhibited resistance with infection type (IT) 0 to a Bgt mixture including E09, E18, E20 and other isolates originally collected from northern China at the adult plant stage. At the seedling stage, the Bgt isolate E09 was used to determine the inheritance of the powdery mildew resistance in DR88. All the 10 tested DR88 plants (resistant parent) were immune with IT 0, while all the 10 tested Mo75 plants (susceptible parent) were highly susceptible with IT 4. All the 30 F1 plants crossed by DR88 and Mo75 also showed immunity, indicating the dominant inheritance of the resistance gene(s) in DR88. The F2 population of 230 plants segregated into 174 resistant (IT 0) and 56 susceptible (IT 4) individuals, fitting a 3:1 ratio for monogenic segregation (χ2 = 0.02; P = 0.88) (Fig. 1a). Further analysis using their derived 105 F2:3 families segregated 25 homozygous resistant, 59 segregating and 21 homozygous susceptible families after inoculating with the Bgt isolate E09, which also fitted the expected ratio of 1:2:1 for monogenic segregation (χ2 = 1.91; P = 0.38). Therefore, the resistance to the Bgt isolate E09 in DR88 is controlled by a single dominant gene, temporarily designated PmDR88. To further validate whether PmDR88 confers resistance to other Bgt isolates, three additional Bgt isolates, E18, E20, and E21, were randomly selected to inoculate the 105 F2:3 families. The phenotypic statistics for each F2:3 family were consistent with the results observed with Bgt isolate E09, indicating that PmDR88 also confer resistance to these Bgt isolates.
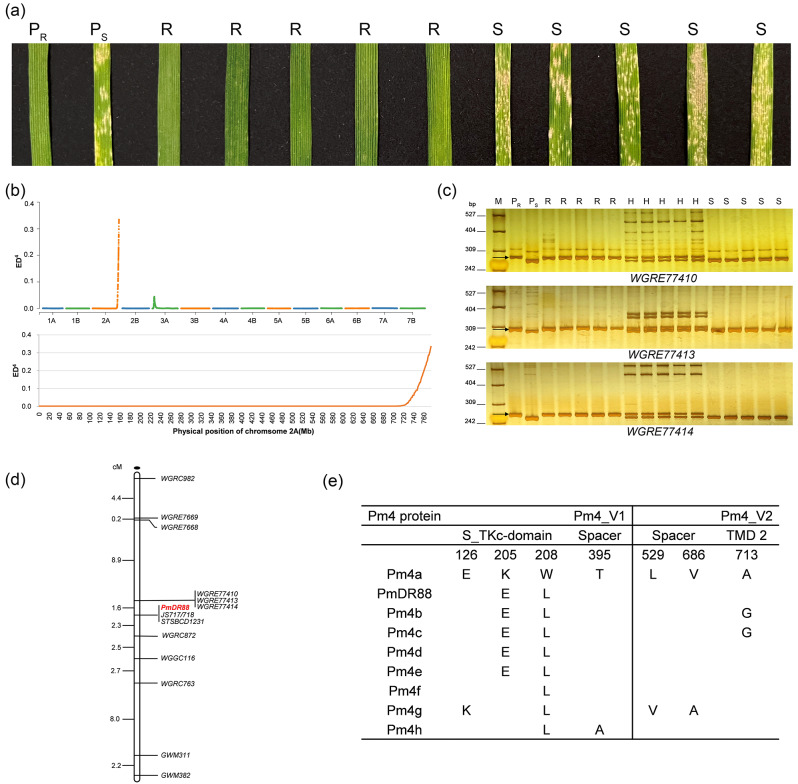
Fig. 1.
Molecular identification and mapping of PmDR88. (a) Reaction patterns of DR88, Mo75, and F2 plants crossed by DR88 and Mo75 to the Blumeria graminis f. sp. tritici isolate E09 at the seeding stage. PR, DR88. PS, Mo75. R, resistant F2 plants. S, susceptible F2 plants. (b) Distribution of the single nucleotide polymorphisms (SNPs) between the resistant and susceptible bulks on 21 chromosomes and chromosome 2A revealed by BSR-seq. (c) PCR amplification patterns of molecular markers WGRE77410, WGRE77413, and WGRE77414 in F2 plants crossed by Mo75 and DR88. M, pBR322/Msp I. PR, DR88. PS, Mo75. R, homozygous resistant F2 plants. H, heterozygous resistant F2 plants. S, homozygous susceptible F2 plants. Arrows indicate the polymorphic DNA bands. (d) Genetic linkage map of PmDR88. (e) Protein sequence comparison of the Pm4 variants, where blank spaces represent identical amino acids to Pm4a
Analysis of candidate interval using bulked segregant RNA-Seq (BSR-Seq)
BSR-Seq generated 201,915,390 and 211,013,912 raw reads from the resistant and susceptible bulks, respectively. After filtering, 91.0% and 91.9% of the reads were qualified for subsequent analysis, with 170,411,994 (84.4%) and 150,648,246 (71.4%) aligned to the durum wheat Svevo reference genome v1 [17]. Single nucleotide polymorphisms (SNP) calling identified 8,864 high-quality SNPs between resistant and susceptible bulks, of which 1,322 were located on chromosome 2A. Using the Euclidean distance (ED) algorithm, a high-confidence candidate interval was identified on chromosome arm 2AL (743–776 Mb) (Fig. 1b).
Mapping of PmDR88
Based on the Insertions/deletions (InDels) variations over 3 bp identified between resistant and susceptible bulks, 9 markers were developed using the PrimerServer function in WheatOmics 1.0 (http://202.194.139.32/), and five of them, including WGRE7668, WGRE7669, WGRE77410, WGRE77413 and WGRE77414, showed consistent polymorphisms between the resistant and susceptible parents as well as bulks (Fig. 1c and Table S1). Additionally, 18 known markers located on the chromosome 2AL which are linked to Pm4 locus [23–32] were also used for genotyping the resistant and susceptible parents and bulks (Table S2). Among them, eight markers, including JS717×JS718, STSBCD1231, WGRC763, WGGC116, GWM311, GWM382, WGRC982 and WGRC872, showed consistent polymorphism between the resistant and susceptible parents and bulks.
To confirm their linkage with PmDR88, the F2 population consisting of 230 plants from the cross of DR88 and Mo75 was genotyped using the 13 markers which showed polymorphisms. The results demonstrated that all these 13 markers were closely linked with PmDR88. Then, a linkage map was constructed, which revealed PmDR88 was flanked by two markers, WGRE77410 and WGRC872, with genetic distances of 1.6 and 2.3 cM, respectively (Fig. 1d). Two markers, JS717×JS718 and STSBCD1231, were co-segregated with PmDR88, with JS717×JS718 being the diagnostic marker of Pm4 [23]. Additionally, the 105 F2:3 families were genotyped, and the results confirmed that JS717×JS718 remained co-segregating with the phenotypes (Fig. S1) Furthermore, to accurately validate the relationship between PmDR88 and JS717×JS718, a large F2 population containing 5,174 plants was genotyped using JS717×JS718. The results indicate that JS717×JS718 still co-segregated with PmDR88, suggesting that PmDR88 should be located at the Pm4 locus.
Haplotype analysis of PmDR88 with Pm4 alleles
The sequence of PmDR88 was isolated through homology-based cloning to dissect its relationship with the reported Pm4 alleles [23]. Sanger sequencing showed that PmDR88_V1 has only a synonymous single nucleotide variant (SNV) c.T1026C, which did not result in a change in the 342th amino acid proline, thereby maintaining consistent amino acid sequence with that of Pm4b_V1 (Table S3). When compared with Pm4b_V2, PmDR88_V2 carries a nonsynonymous SNV c.G2138C in the phosphoribosyl transferase C-terminal domain, leading to the 713th amino acid change from glycine to alanine (Table S3). Meanwhile, PmDR88_V2 possesses a synonymous SNV c.G2091C without altering the 697th amino acid valine. Furthermore, when compared with other Pm4 alleles, the amino acid sequences of PmDR88 were identical to Pm4d and Pm4e (Fig. 1e). These findings suggest that PmDR88 could represent Pm4d at the level of amino acid sequence.
Expression analysis of PmDR88 with Pm4 alleles
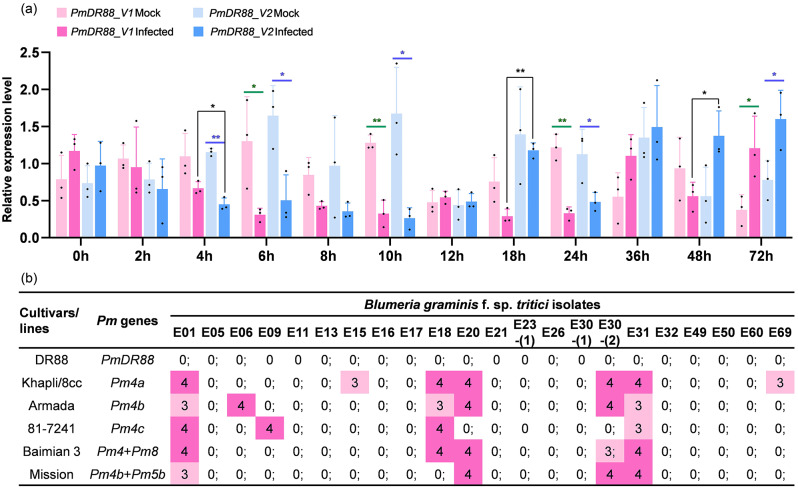
Compared with the mock-inoculated plants, both PmDR88_V1 and PmDR88_V2 were down-regulated at 0–24 h post inoculation (hpi) after Bgt inoculation, with significant differences between them observed at 4 and 18 hpi. At 36 hpi, both transcripts were up-regulated, showing contrasting expression patterns at 48 hpi. At 72 hpi, both transcripts exhibited significantly higher expression levels compared with mock-inoculated plants. These results suggest that PmDR88_V1 and PmDR88_V2 display similar expression patterns post-inoculation, with notable differences at specific time points (Fig. 2a).
Fig. 2.
Powdery mildew resistance pattern analysis of PmDR88. (a) Expression patterns of the PmDR88_V1 and PmDR88_V2 splice variants in mock-inoculated or Bgt-inoculated durum wheat accession DR88 plants during a 72-hour time course. Error bars represent SD based on three biological replicates. Statistical analysis was done using t-test, * and ** indicate significant (P < 0.05) and extremely significant (P < 0.01), respectively. The black dots indicate biological replicates. (b) Comparison of seedling responses of DR88 and several carriers of Pm4 alleles to 22 Blumeria graminis f. sp. tritici isolates. Responses were recorded as infection types at the seedling stage using a 0–4 scale [56], in which 0–2 were considered resistant and 3–4 susceptible
Resistant spectrum analysis of PmDR88 with the reported Pm4 alleles
To analyze the resistance ability of PmDR88 and evaluate its breeding value, 22 Bgt isolates were used to compare the reaction patterns of DR88 and 44 donors with reported Pm genes, including Khapli/8 cc carrying Pm4a, Armada carrying Pm4b, 81-7241 carrying Pm4c, Baimian 3 carrying Pm4 and Pm8, Mission carrying Pm4b and Pm5b. The results showed that PmDR88 was highly resistant to all the tested Bgt isolates (IT 0–0;). In comparison, other donors containing Pm4 alleles all showed narrowed and weaker resistant spectra, and were susceptible to at least four Bgt isolates (Fig. 2b and Table S4). Therefore, PmDR88 have high breeding value against powdery mildew.
Molecular markers available for MAS
To facilitate MAS for PmDR88, the diagnostic marker JS717×JS718 was tested across 24 susceptible wheat cultivars/breeding lines. JS717×JS718 successfully yielded same-size amplicons in DR88 as reported in Sánchez-Martín et al. [23] but not in the 24 susceptible cultivars/breeding lines, indicating its utility in MAS. Among the three closely linked markers, only WGRE77410 produced amplicons of different sizes in DR88 and in 21 of the 24 susceptible wheat cultivars/breeding lines, with the exceptions of Zhongmai 5215, Lunxuan 49, and Lunxuan 69. (Fig. S2 and Table S5). Conversely, both WGRE77413 and WGRE77414 failed to produce polymorphic bands between DR88 and 24 susceptible wheat cultivars/breeding lines without Pm4 alleles, indicating their unsuitability for MAS tracking of PmDR88. Therefore, the dominant co-segregating marker JS717×JS718 and co-dominant closely-linked marker WGRE77410 can be effectively employed in MAS to track PmDR88.
Discussion
In this study, the durum wheat accession DR88 exhibited high levels of resistance to powdery mildew during whole growth period. A dominant Pm gene, PmDR88, was identified as responsible for the resistance to powdery mildew in DR88. Using BSR-Seq and molecular marker analysis, we mapped PmDR88 in the Pm4 locus, which has developed multiple alleles from various donors, including Pm4a from T. dicoccum [33], Pm4b from T. carthlicumn [34], Pm4c and Pm4e from common wheat [28, 31] and Pm4d from T. monococcum [27]. Cloning of the Pm4 alleles revealed that Pm4 encodes a putative chimeric protein consisting of a serine/threonine kinase and multiple C2 domains and transmembrane regions [23]. Notably, constitutive alternative splicing of this gene produces two isoforms, Pm4b_V1 and Pm4b_V2, each essential for resistance and differing in protein domain topologies [23]. Further investigation using the diagnostic marker JS717×JS718 identified new susceptible alleles, Pm4f and Pm4g, as well as a resistant allele, Pm4h [23]. Besides these alleles mentioned above, several genes have been reported as allelic or closely linked to Pm4, such as PmPS5A [35], pmLK906 [36], PmHNK54 [37], Pm50 [38], pmXMM [32], PmYAV [39] and PmGR-18 [40]. Our findings revealed that PmDR88 could represents Pm4d at the level of amino acid sequence, which significantly enhances and broadens resistance to powdery mildew. However, we cannot rule out the possibility of synonymous mutations at the nucleotide level. Further comparisons of the nucleotide sequences among Pm4d carriers will be necessary in future studies.
In previous studies, multiple alleles with diverse sequences and resistance patterns during evolution have been observed at several Pm loci, including Pm1 [41], Pm2 [42], Pm3 [43], Pm4 [23], Pm5 [44], Pm55 [45] and Pm60 [46]. For instance, over 10 alleles at the Pm60 locus exhibit distinct resistant spectra, suggesting a potential co-evolution relationship between the resistant gene and Bgt isolates [14]. Specific interactions with inhibitors among different Pm55 alleles result in developmental-stage and tissue-specific resistance to powdery mildew [44]. In this study, DR88 showed resistance to all 22 tested Bgt isolates, demonstrating a broader resistance spectrum. Even when compared with the other 39 tested Pm gene donors in this study, DR88 shows exceptional resistance, comparable to some of the most robust Pm genes, such as Pm1c, Pm12, Pm13, Pm16, Pm21, Pm24, Pm36 and Pm40. Additionally, DR88 showed improved resistance compared with synthetic hexaploid wheat YAV249, which carries the new Pm4 variant PmYAV [39]. The wheat breeding line GR-18, carrying PmGR-18 [40] and the Chinese landrace Xiaomaomai, carrying pmXMM [32], both confirmed to be Pm4d, were found to be resistant to 5 of 10 and 12 of 40 tested Bgt isolates, respectively, in their respective studies. As described in previous study [23], Pm4d-containing genotypes exhibited similar response patterns to Pm4b-containing genotypes. Therefore, DR88 and PmDR88 should be valuable germplasm and gene resources in wheat powdery mildew resistance breeding.
We analyzed the expression patterns of PmDR88 transcripts following Bgt infection and compared them with those of other reported Pm4 alleles. Previous studies have shown that the transcripts of Pm4b (Pm4b_V1 and Pm4b_V2) contribute similarly to Bgt infection in the Federation background [23]. PmYAV transcripts (PmYAV_V1 and PmYAV_V2) display similar expression patterns, though up-regulation is more pronounced in PmYAV_V2, suggesting a higher contribution of PmYAV_V2 to resistance [39]. The two transcripts of PmGR-18, identified as Pm4d, show similar expression patterns and levels from 0 to 16 hpi, but with PmGR18_V2 exhibiting significantly lower expression levels during the period from 20 to 72 hpi [40]. In DR88, PmDR88_V1 and PmDR88_V2 display similar expression patterns except at 48 dpi. Additionally, significant differences between them are observed at 4, 18 and 48 dpi (Fig. 2b). Therefore, PmDR88 has a distinctive expression profile compared with the reported Pm4 alleles. We speculate that several factors may account for this enhanced and distinct resistance: (1) different alternative splicing patterns may lead to alterations in protein structure and protein-protein interactions, thereby impacting biological functions or modulating the synergistic effects between the two proteins [47, 48], (2) variations in genetic background may influence resistance activities, and (3) the purity of Bgt isolates and differences in their virulent spectrum may affect transcript expression levels.
Looking ahead, our ultimate goal is to incorporate PmDR88 into breeding programs, leveraging its strong resistance profile. Pm4 alleles encode a novel chimeric protein rather than a typical NLR protein. These alleles possess diverse immune receptors and resistance mechanisms, making them highly valuable for breeding [49, 50]. Extensive breeding practices have proved that Pm4 is one of the most widely used Pm genes in wheat production, suggesting it is an elite locus without negative effect on breeding outcomes [26, 51, 52]. For PmDR88, its broader resistant spectrum further endowed higher breeding potential against powdery mildew. To efficiently transfer PmDR88, MAS remains the most efficient approach for transferring PmDR88 into wheat backgrounds. To extend its effective lifespan, pyramiding PmDR88 with other Pm genes (especially those encoding different classes of proteins, such as NLR proteins) to develop durable resistance is the preferred strategy. For instance, Pm3 alleles-pyramided lines showed strongly improved powdery mildew resistance in the field compared with their parental lines [53]. During the transfer of PmDR88, the co-segregating marker JS717×JS718, located at the Pm4 locus, can accurately trace Pm4 alleles, including PmDR88. However, it is a dominant marker incapable of distinguishing between homozygous and heterozygous plants. To overcome this limitation, we developed a new co-dominant marker WGRE77410 closely-linked with PmDR88 that can be used in MAS. By combining these two markers, PmDR88 has been transferred into more than 10 susceptible commercial wheat cultivars without Pm4 alleles through conventional hybridization and MAS strategy, including Shixin 6171, Jimai 330, Jimai 340, Henong 826, Zhoumai 11, etc. We are now in the process of selecting stable progenies that exhibit both high levels of resistance to powdery mildew and favorable yield and agronomic traits.
Conclusion
We identified an allele of the powdery mildew resistance gene Pm4, designated as PmDR88, in the durum wheat accession DR88. Through BSR-seq analysis and gene mapping, PmDR88 was localized within the Pm4 locus on chromosome arm 2AL, and could represents Pm4d at the level of amino acid sequence. In spite of this, PmDR88 exhibits distinct expression patterns, suggesting potential differences in its resistance mechanism. Resistance spectrum analysis demonstrated that DR88 confers broad and high levels resistance to various Bgt isolates, surpassing other tested carriers with known Pm4 alleles. Two molecular markers JS717×JS718 (dominant) and WGRE77410 (co-dominant) were selected, which will facilitate the efficient transfer of PmDR88 in wheat breeding through MAS. Our findings provide valuable genetic resources and tools for wheat disease resistance breeding.
Materials and methods
Plant materials
The durum wheat accessions DR88 (highly resistant to powdery mildew), kindly provided by Prof. Lihui Li (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences), and Mo75 (highly susceptible to powdery mildew), kindly provided by Prof. Zhiyong Liu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), were crossed to generate F1, F2 and F2:3 progenies for identifying powdery mildew resistance gene(s). Forty-four wheat genotypes (Table S4) with known Pm genes were collected for comparative reaction pattern analysis with DR88 in the greenhouse at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences. The susceptible wheat cultivar Heng 4399 served as both a control and Bgt inoculum spreader. Additionally, 24 cultivars or breeding lines from China (Table S5), maintained in our laboratory, were selected for assessing polymorphism in gene-linked DNA markers.
Phenotypic evaluation and genetic analysis
To evaluate powdery mildew resistance at the adult stage, DR88 was inoculated with a Bgt isolate mixture including isolates E09, E18, and E20 and other isolates originally collected from northern China at Luancheng, Hebei, China (37°50′N,114°40′E), during 2020–2022 cropping seasons. DR88 was planted in three replicated plots organized in a randomized complete block design, with each plot comprising three rows measuring 1.5 m in length with a 20 cm inter-row spacing. Heng 4399 was planted around the plots to enhance Bgt spread. Disease reactions were evaluated after the full heading stage [54] using a 0–9 scale, where 0–4 indicated resistance and 5–9 indicated susceptibility [55]. Assessments were performed twice, with a one-week interval.
Twenty-two single-pustule-derived Bgt isolates were utilized to evaluate seedling resistance of DR88 and compare reaction patterns with the 44 documented wheat genotypes (Table S4). Independent inoculation was conducted at the one-leaf stage, and seedlings were assessed 10–14 days post-inoculation using a 0–4 scale [56], where 0, 0;, 1, and 2 were classified as resistant, and 3 and 4 as susceptible. Three independent replications were included.
Inheritance of resistance in DR88 was firstly examined using Bgt isolate E09, with phenotypic reactions across DR88, Mo75, and their F1, F2 and F2:3 generations. If the segregation ratio against isolate E09 was consistent with that of monogenetic inheritance, two other isolates avirulent on DR88 will be further used to inoculate the F2:3 generations. The phenotypic reaction of the F2 generations was confirmed by testing each of the F2:3 families with 30 seeds. Chi-squared (χ2) test was applied to test deviations between observed phenotypic data and theoretically expected segregation ratios using SPSS 16.0 (SPSS Inc, Chicago, IL, USA). Values for significance at P = 0.05 are 3.84 (1 df) and 5.99 (2 df).
BSR-Seq analysis
Thirty resistant (IT0, balanced mix for each plant) and susceptible (IT4, balanced mix for each plant) F2 plants were selected to isolate respective total RNA from their primary leaves, respectively. cDNA libraries for the resistant and susceptible bulks were constructed using Stranded mRNA LTSample Prep Kits (Illumina, San Diego, CA, USA). RNA-Seq of eligible cDNA libraries was conducted on the DNBSEQ-T7 platform at Chengdu Tiancheng Future Technology Co., Ltd. (Chengdu, China), with a target output of 20 Gb clean data for each bulk. After sequencing the cDNA libraries, the raw data were processed using Fastp software (v0.12.4) to remove reads containing adapters, poor quality, and N proportion ≥ 10 [57]. High-quality clean data was obtained and aligned to the durum wheat Svevo reference genome v1 [17] using BWA software (v0.7.17). Samtools (v.1.9) was used to sort the reads and remove duplicates. Then, SNPs were identified with GATK (v4.1.3.0) (https://gatk.broadinstitute.org/hc/en-us). The quality of the SNP was assessed using the Bcftools (v1.9) software following the thresholds of QUAL > 30 and DP ≥ 5 [58]. The ED algorithm was used to refine candidate intervals [59]. Candidate intervals with higher confidence (95%) and SNPs with larger than the threshold ΔSNP index value (set as 0.5) were regarded as candidate loci associated with powdery mildew resistance.
Marker development and gene mapping
InDels variations over 3 bp identified between resistant and susceptible bulks were used to design InDel markers via PrimerServer (http://202.194.139.32/PrimerServer/). In addition, 18 previously documented markers were also used for marker screening (Table S2). Polymorphisms were detected between resistant and susceptible parents and bulks, and the resulting markers were subsequently genotyped on the F2 population of DR88 × Mo75 for gene mapping. Co-segregating markers were further genotyped on a large population of over 5000 F2 plants for precise locus identification. PCR amplification system, PCR reaction system and gel visualization were carried out described by Han et al. [60]. When the genotyping data on the molecular marker analysis and phenotyping data from the evaluation of disease resistance were obtained, the linkage map was constructed using MAPMAKER 3.0 with the Kosambi mapping function as previously reported [61, 62].
Cloning and sequence analysis
Considering the Pm gene in DR88 mapped in the Pm4 locus, homologous sequences of the Pm4 gene were isolated from the cDNA of DR88 using nested PCR with the reported primers GH398×GH399, GH400×GH401, GH398×GH407 and GH407×GH400 [23]. The sequences, alternative splicing patterns and their encoded amino acid sequence were compared with those of reported Pm4 alleles.
RNA extraction and qRT-PCR
qRT-PCR was carried out to profile the expression of PmDR88_V1 and PmDR88_V2 after inoculation with Bgt isolate E09. Young leaves of DR88 were sampled at 0, 2, 4, 6, 8, 10, 12, 18, 24, 36, 48 and 72 h post inoculation (hpi) with Bgt isolate E09. At each point above, the young leaves of DR88 without any inoculation were also sampled as mock-infection. Total RNA was extracted using the TRIzol reagent (TaKaRa, Kyoto, Japan) and converted to cDNA using FastQuantRT Kit (Tiangen, Beijing, China). qRT-PCR was performed using SYBR Green (TaKaRa, Kyoto, Japan) on a Bio-Rad CFX real-time PCR system (Bio-Rad, Hercules, CA), with TaActin as an internal control [63]. Gene expression was calculated using the 2−ΔΔCT method [64], with three technical replicates for each of three biological replicates. Primers used in this study were listed in Table S2.
Identification of the markers for MAS
To transfer the target Pm gene in DR88, co-segregating and closely linked markers were used to genotype DR88 and 24 powdery mildew-susceptible wheat cultivars/breeding lines sourced from various regions across China. Markers showing polymorphism between DR88 and susceptible genotypes were deemed suitable for MAS in these genetic backgrounds.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1: Additional file 1: Table S1: Primers developed in this study. Table S2: Primers used in this study. Table S3: cDNA sequences of PmDR88_V1 and PmDR88_V2. Table S4: Responses of DR88 and 44 wheat accessions with known powdery mildew resistance genes to 22 different Blumeria graminis f. sp. tritici isolates. Table S5: Detection results of molecular markers JS717xJS718 and WGRE77410 for PmDR88 in 24 cultivars or breeding lines from China.
Supplementary Material 2: Additional file 2: Fig. S1: PCR amplification patterns of the molecular marker JS717xJS718 in 105 F2:3 families from the cross of DR88 and Mo75. (a) M, Trans2K Plus II. 1-84, 25 homozygous resistant and 59 segregating families after inoculating with the Blumeria graminis f. sp. tritici isolate E09. 85, Mo75. 86, DR88. (b) M, Trans2K Plus II. 1-23, 21 homozygous susceptible families after inoculating with the Bgt isolate E09. 22, Mo75. 23, DR88.
Supplementary Material 3: Fig. S2: PCR amplification patterns of molecular markers JS717xJS718 and WGRE77410 in various wheat cultivars/lines from China. M, pBR322/Msp I for JS717xJS718 and Trans2K Plus II for WGRE77410. 1, DR88. 2, Sinxin 6171. 3, Jimai 330. 4, Jimai 340. 5, Henong 826. 6, Hanmai 4. 7, Yumai 35. 8, Yumai 51. 9, Fanyumai 18. 10, Fanyumai 20. 11, Xinong 9718. 12, Zhongmai 5215. 13, Lunxuan 49. 14, Lunxuan 69.
Acknowledgements
The authors express their sincere gratitude to Prof. Lihui Li (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China) for providing the DR88 seeds and to Prof. Zhiyong Liu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China) for providing the Mo75 seeds.
Author contributions
DA and Y Zhang conceived and supervised the study. GH analyzed the data and wrote the manuscript. LX performed the molecular experiments. TG constructed the genetic populations. LX, TG, YJ, FS, HY and JW performed the field experiments. GH, SZ and ZS collected the data. GH, LX, Y Zhou and WL evaluated the phenotype. DA revised the writing of the article. All authors have approved the final manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (32272105) and the National Key Research and Development Program of China (2021YFD1200600). The funding bodies were not involved in the design of the study, and collection, analysis, interpretation of data and manuscript writing.
Data availability
All the data generated or analyzed during the current study were included in the manuscript and its additional files. Sequence data were deposited at NCBI GenBank under the accession numbers PQ550686 (PmDR88_V1 CDS) and PQ550687 (PmDR88_V2 CDS). The raw data and the Bgt isolates used in this study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yelun Zhang, Email: zhang-ye-lun@126.com.
Diaoguo An, Email: dgan@sjziam.ac.cn.
References
- 1.Shewry PR, Hey SJ. The contribution of wheat to human diet and health. Food Energy Secur. 2015;4(3):178–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun H, Hu J, Song W, Qiu D, Cui L, Wu P, Zhang H, Liu H, Yang L, Qu Y, Li Y, Li T, Cheng W, Zhou Y, Liu Z, Li J, Li H. Pm61: a recessive gene for resistance to powdery mildew in wheat landrace Xuxusanyuehuang identified by comparative genomics analysis. Theor Appl Genet. 2018;131(10):2085–97. [DOI] [PubMed] [Google Scholar]
- 3.Jin Y, Han G, Zhang W, Bu B, Zhao Y, Wang J, Liu R, Yang H, Xu H, Ma P. Evaluation and genetic dissection of the powdery mildew resistance in 558 wheat accessions. New Crops. 2024;1:100018. [Google Scholar]
- 4.Han G, Yan H, Wang J, Gu T, Cao L, Liu S, Li X, Zhou Y, Fan J, Shi Z, Liu H, Li L, An D. Development and identification of two novel wheat-rye 6R derivative lines with adult-plant resistance to powdery mildew and high-yielding potential. Crop J. 2024;12(1):308–13. [Google Scholar]
- 5.Wang B, Meng T, Xiao B, Yu TY, Yue TY, Jin YL, Ma PT. Fighting wheat powdery mildew: from genes to fields. Theor Appl Genet. 2023;136(9):196. [DOI] [PubMed] [Google Scholar]
- 6.He HG, Chen ZZ, Fan RC, Zhang J, Zhu SY, Wang JL, Zhang QY, Gao AL, Gong SJ, Zhang L, Li YN, Zhao YT, Krattinger SG, Shen QH, Li HJ, Wang YJ. A kinase fusion protein from Aegilops longissima confers resistance to wheat powdery mildew. Nat Commun. 2024;15(1):6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li HH, Men WQ, Ma C, Liu QW, Dong ZJ, Tian XB, Wang CL, Liu C, Gill HS, Ma PT, Zhang ZB, Liu B, Zhao Y, Sehgal SK, Liu WX. Wheat powdery mildew resistance gene Pm13 encodes a mixed lineage kinase domain-like protein. Nat Commun. 2024;15(1):2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li MM, Zhang HZ, Xiao HX, Zhu KY, Shi WQ, Zhang D, Wang Y, Yang LJ, Wu QH, Xie JZ, Chen YQ, Qiu D, Guo GH, Lu P, Li BB, Dong L, Li WL, Cui XJ, Li LC, Tian XB, Yuan CG, Li YW, Yu DZ, Nevo E, Fahima T, Li HJ, Dong LL, Zhao YZ, Liu ZY. A membrane associated tandem kinase from wild emmer wheat confers broad-spectrum resistance to powdery mildew. Nat Commun. 2024;15(1):3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li YH, Wei ZZ, Sela H, Govta L, Klymiuk V, Roychowdhury R, Chawla HS, Ens J, Wiebe K, Bocharova V, Ben-David R, Pawar PB, Zhang Y, Jaiwar S, Molnár I, Doležel J, Coaker G, Pozniak CJ, Fahima T. Dissection of a rapidly evolving wheat resistance gene cluster by long-read genome sequencing accelerated the cloning of Pm69. Plant Commun. 2024;5(1):100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Dong ZJ, Miao JN, Liu QW, Ma C, Tian XB, He JQ, Bi HH, Yao W, Li T, Gill HS, Zhang ZB, Cao AZ, Liu B, Li HH, Sehgal SK, Liu WX. Pm57 from Aegilops searsii encodes a tandem kinase protein and confers wheat powdery mildew resistance. Nat Commun. 2024;15(1):4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan C, Li G, Cowger C, Carver BF, Xu X. Characterization of Pm59, a novel powdery mildew resistance gene in Afghanistan wheat landrace PI 181356. Theor Appl Genet. 2018;131(5):1145–52. [DOI] [PubMed] [Google Scholar]
- 12.Han G, Wang J, Yan H, Cao L, Liu S, Li X, Zhou Y, Liu W, Gu T, Shi Z, Liu H, Li L, An D. Development and molecular cytogenetic identification of a new wheat-rye 6RL ditelosomic addition and 1R (1B) substitution line with powdery mildew resistance. J Integr Agric. 2023. 10.1016/j.jia.2023.10.004. In Press. [Google Scholar]
- 13.Wicker T, Oberhaensli S, Parlange F, Buchmann JP, Shatalina M, Roffler S, Ben-David R, Doležel J, Šimková H, Schulze-Lefert P, Spanu PD, Bruggmann R, Amselem J, Quesneville H, van Ver E, Paape T, Shimizu KK, Keller B. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat Genet. 2013;45(9):1092–6. [DOI] [PubMed]
- 14.Wei WX, Liu NN, Zhang SN, Zhang J, Pan W, Xie XM, Yang ZH, Sun JN, Ma J, Hu ZR, Guo WL, Luo QL, Xie JZ, He F, Li YH, Xie CJ, Sun QX. Mapping of powdery mildew resistance genes transferred to common wheat from wild emmer wheat revealed three functional Pm60 haplotypes. Crop J. 2024;12(2):540–8. [Google Scholar]
- 15.Xiao J, Liu B, Yao Y, Guo Z, Jia H, Kong L, Zhang A, Ma W, Ni Z, Xu S, Lu F, Jiao Y, Yang W, Lin X, Sun S, Lu Z, Gao L, Zhao G, Cao S, Chen Q, Zhang K, Wang M, Wang M, Hu Z, Guo W, Li G, Ma X, Li J, Han F, Fu X, Ma Z, Wang D, Zhang X, Ling HQ, Xia G, Tong Y, Liu Z, He Z, Jia J, Chong K. Wheat genomic study for genetic improvement of traits in China. Sci China Life Sci. 2022;65(9):1718–75. [DOI] [PubMed] [Google Scholar]
- 16.Qian Z, Han G, Yu N, Liu C, Han R, Jameson P, Wang J, Zhao Y, Xiao B, Liu R. Fine mapping of the powdery mildew resistance gene PmXQ-0508 in bread wheat. Crop J. 2024;12:1176–84. [Google Scholar]
- 17.Maccaferri M, Harris NS, Twardziok SO, Pasam RK, Gundlach H, Spannagl M, Ormanbekova D, Lux T, Prade VM, Milner SG, Himmelbach A, Mascher M, Bagnaresi P, Faccioli P, Cozzi P, Lauria M, Lazzari B, Stella A, Manconi A, Gnocchi M, Moscatelli M, Avni R, Deek J, Biyiklioglu S, Frascaroli E, Corneti S, Salvi S, Sonnante G, Desiderio F, Marè C, Crosatti C, Mica E, Özkan H, Kilian B, De Vita P, Marone D, Joukhadar R, Mazzucotelli E, Nigro D, Gadaleta A, Chao S, Faris JD, Melo ATO, Pumphrey M, Pecchioni N, Milanesi L, Wiebe K, Ens J, MacLachlan RP, Clarke JM, Sharpe AG, Koh CS, Liang KYH, Taylor GJ, Knox R, Budak H, Mastrangelo AM, Xu SS, Stein N, Hale I, Distelfeld A, Hayden MJ, Tuberosa R, Walkowiak S, Mayer KFX, Ceriotti A, Pozniak CJ, Cattivelli L. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet. 2019;51(5):885–95. [DOI] [PubMed] [Google Scholar]
- 18.Pont C, Leroy T, Seidel M, Tondelli A, Duchemin W, Armisen D, Lang D, Bustos–Korts D, Goué N, Balfourier F, Molnár–Láng M, Lage J, Kilian B, Özkan H, Waite D, Dyer S, Letellier T, Alaux M; Wheat and Barley Legacy for Breeding Improvement (WHEALBI) consortium;, Russell J, Keller B, van Eeuwijk F, Spannagl M, Mayer KFX, Waugh R, Stein N, Cattivelli L, Haberer G, Charmet G, Salse J. Tracing the ancestry of modern bread wheats. Nat Genet. 2019;51(5):905–11. [DOI] [PubMed]
- 19.Bennett FG. Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes. Plant Pathol. 1984;33(3):279–300. [Google Scholar]
- 20.Huang X, Hsam SL, Mohler V, Röder MS, Zeller FJ. Genetic mapping of three alleles at the Pm3 locus conferring powdery mildew resistance in common wheat (Triticum aestivum L). Genome. 2004;47(6):1130–6. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z, Kong X, Zhou R, Jia J. Identification and microsatellite markers of a resistance gene to powdery mildew in common wheat introgressed from Triticum durum. Acta Bot Sin. 2004;46(7):867–72. [Google Scholar]
- 22.He H, Liu R, Ma P, Du H, Zhang H, Wu Q, Yang L, Gong S, Liu T, Huo N, Gu YQ, Zhu S. Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor Appl Genet. 2021;134(1):53–62. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Martín J, Widrig V, Herren G, Wicker T, Zbinden H, Gronnier J, Spörri L, Praz CR, Heuberger M, Kolodziej MC, Isaksson J, Steuernagel B, Karafiátová M, Doležel J, Zipfel C, Keller B. Wheat Pm4 resistance to powdery mildew is controlled by alternative splice variants encoding chimeric proteins. Nat Plants. 2021;7(3):327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma ZQ, Wei JB, Cheng SH. PCR-based markers for the powdery mildew resistance gene Pm4a in wheat. Theor Appl Genet. 2004;109(1):140–5. [DOI] [PubMed] [Google Scholar]
- 25.Ullah KN, Li N, Shen T, Wang P, Tang W, Ma S, Zhang Z, Jia H, Kong Z, Ma Z. Fine mapping of powdery mildew resistance gene Pm4e in bread wheat (Triticum aestivum L). Planta. 2018;248(5):1319–28. [DOI] [PubMed] [Google Scholar]
- 26.Wu P, Xie J, Hu J, Qiu D, Liu Z, Li J, Li M, Zhang H, Yang L, Liu H, Zhou Y, Zhang Z, Li H. Development of molecular markers linked to powdery mildew resistance gene Pm4b by combining SNP discovery from transcriptome sequencing data with bulked segregant analysis (BSR-Seq) in wheat. Front Plant Sci. 2018;9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmolke M, Mohler V, Hartl L, Zeller FJ, Hsam SLK. A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol Breed. 2012;29:449–56. [Google Scholar]
- 28.Li N, Jia H, Kong Z, Tang W, Ding Y, Liang J, Ma H, Ma Z. Identification and marker-assisted transfer of a new powdery mildew resistance gene at the Pm4 locus in common wheat. Mol Breed. 2017;37:79. [Google Scholar]
- 29.Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149(4):2007–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi YJ, Liu HY, Huang XQ, An LZ, Wang F, Wang XL. Development of molecular markers linked to the wheat powdery mildew resistance gene Pm4b and marker validation for molecular breeding. Plant Breeding. 2008;127(2):116–20. [Google Scholar]
- 31.Hao Y, Liu A, Wang Y, Feng D, Gao J, Li X, Liu S, Wang H. Pm23: a new allele of Pm4 located on chromosome 2AL in wheat. Theor Appl Genet. 2008;117(8):1205–12. [DOI] [PubMed] [Google Scholar]
- 32.Yao D, Ijaz W, Liu Y, Hu J, Peng W, Zhang B, Wen X, Wang J, Qiu D, Li H, Xiao S, Sun G. Identification of a Pm4 allele as a powdery mildew resistance gene in wheat line Xiaomaomai. Int J Mol Sci. 2022;23(3):1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briggle LW. Transfer of resistance to Erysiphe Graminis f. sp. tritici from Khapli Emmer and Yuma durum to hexaploid wheat. Crop Sci. 1966;6(5):459–61. [Google Scholar]
- 34.The TT, McIntosh RA, Bennett FGA. Cytogenetical studies in wheat. IX.* Monosomic analyses, telocentric mapping and linkage relationships of genes Sr21, Pm4 and Mle. Aust J Biol Sci. 1979;32(1):115–25.
- 35.Zhu Z, Zhou R, Jia J. Identification of powdery mildew resistance genes in advanced wheat lines using molecular markers. Acta Agron Sin. 2005;31(8):977–82. [Google Scholar]
- 36.Niu J, Jia H, Jun Y, Wang B, Shen T. Development of an STS marker linked to powdery mildew resistance genes PmLK906 and Pm4a by gene chip hybridization. J Integr Agric. 2010;9(3):331–6. [Google Scholar]
- 37.Xu W, Li C, Hu L, Wang H, Dong H, Zhang J, Zan X. Identification and molecular mapping of PmHNK54: a novel powdery mildew resistance gene in common wheat. Plant Breed. 2011;130(6):603–7. [Google Scholar]
- 38.Mohler V, Bauer C, Schweizer G, Kempf H, Hartl L. Pm50: a new powdery mildew resistance gene in common wheat derived from cultivated emmer. J Appl Genet. 2013;54(3):259–63. [DOI] [PubMed] [Google Scholar]
- 39.Jin Y, Gu T, Li X, Liu H, Han G, Shi Z, Zhou Y, Fan J, Wang J, Liu W, Zhao H, An D. Characterization of a new splicing variant of powdery mildew resistance gene Pm4 in synthetic hexaploid wheat YAV249. Front Plant Sci. 2022;13:1048252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su F, Han G, Yu Z, Liu C, Li H, Wang X, Chang L, Xiao L, Mu Y, Bian Q, Wang F, Jin Y, Ma P. Identification of a Pm4 allele conferring powdery mildew resistance in wheat breeding line GR18-1. Plant Dis. 2023;107(7):2104–11. [DOI] [PubMed] [Google Scholar]
- 41.Hewitt T, Müller MC, Molnár I, Mascher M, Holušová K, Šimková H, Kunz L, Zhang J, Li J, Bhatt D, Sharma R, Schudel S, Yu G, Steuernagel B, Periyannan S, Wulff B, Ayliffe M, McIntosh R, Keller B, Lagudah E, Zhang P. A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 2021;229(5):2812–26. [DOI] [PMC free article] [PubMed]
- 42.Manser B, Koller T, Praz CR, Roulin AC, Zbinden H, Arora S, Steuernagel B, Wulff BBH, Keller B, Sánchez-Martín J. Identification of specificity-defining amino acids of the wheat immune receptor Pm2 and powdery mildew effector AvrPm2. Plant J. 2021;106(4):993–1007. [DOI] [PubMed] [Google Scholar]
- 43.Srichumpa P, Brunner S, Keller B, Yahiaoui N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 2005;139(2):885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie J, Guo G, Wang Y, Hu T, Wang L, Li J, Qiu D, Li Y, Wu Q, Lu P, Chen Y, Dong L, Li M, Zhang H, Zhang P, Zhu K, Li B, Deal KR, Huo N, Zhang Y, Luo MC, Liu S, Gu YQ, Li H, Liu Z. A rare single nucleotide variant in Pm5e confers powdery mildew resistance in common wheat. New Phytol. 2020;228(3):1011–26. [DOI] [PubMed] [Google Scholar]
- 45.Lu C, Du J, Chen H, Gong S, Jin Y, Meng X, Zhang T, Fu B, Molnár I, Holušová K, Said M, Xing L, Kong L, Doležel J, Li G, Wu J, Chen P, Cao A, Zhang R. Wheat Pm55 alleles exhibit distinct interactions with an inhibitor to cause different powdery mildew resistance. Nat Commun. 2024;15(1):503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou S, Wang H, Li Y, Kong Z, Tang D. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2018;218(1):298–309. [DOI] [PubMed] [Google Scholar]
- 47.Syed NH, Kalyna M, Marquez Y, Barta A, Brown JW. Alternative splicing in plants––coming of age. Trends Plant Sci. 2012;17(10):616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy AS, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25(10):3657–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sánchez-Martín J, Keller B. NLR immune receptors and diverse types of non-NLR proteins control race-specific resistance in Triticeae. Curr Opin Plant Biol. 2021;62:102053. [DOI] [PubMed] [Google Scholar]
- 50.O’Hara T, Steed A, Goddard R, Gaurav K, Arora S, Quiroz-Chávez J, Ramírez-González R, Badgami R, Gilbert D, Sánchez-Martín J, Wingen L, Feng C, Jiang M, Cheng S, Dreisigacker S, Keller B, Wulff BBH, Uauy C, Nicholson P. The wheat powdery mildew resistance gene Pm4 also confers resistance to wheat blast. Nat Plants. 2024;10(6):984–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parks R, Carbone I, Murphy JP, Marshall D, Cowger C. Virulence structure of the eastern U.S. wheat powdery mildew population. Plant Dis. 2008;92(7):1074–82. [DOI] [PubMed] [Google Scholar]
- 52.Zeng F, Yang L, Gong S, Gong S, Shi W, Zhang X, Wang H, Xiang L, Xue M, Yu D. Virulence and diversity of Blumeria Graminis f. sp. tritici populations in China. J Integr Agric. 2014;13(11):2424–37. [Google Scholar]
- 53.Koller T, Brunner S, Herren G, Hurni S, Keller B. Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor Appl Genet. 2018;131:861–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zadoks JC, Chang TT, Konzak CF. A decimal code for the growth stages of cereals. Weed Res. 1974;14(6):415–21. [Google Scholar]
- 55.Sheng B, Duan X. Improvement of scale 0–9 method for scoring adult plant resistance to powdery mildew of wheat. Beijing Agr Sci. 1991;1:38–9. [Google Scholar]
- 56.Si Q, Zhang X, Duan X, Sheng B, Zhou Y. On gene analysis and classification of powdery mildew (Erysiphe Graminis f. sp. tritici) resistant wheat varieties. Acta Phytopathol Sin. 1992;22(4):349–55. [Google Scholar]
- 57.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Narasimhan V, Danecek P, Scally A, Xue Y, Tyler-Smith C, Durbin R. BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics. 2016;32(11):1749–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hill JT, Demarest BL, Bisgrove BW, Gorsi B, Su YC, Yost HJ. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013;23(4):687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han G, Yan H, Gu T, Cao L, Zhou Y, Liu W, Liu D, An D. Identification of a wheat powdery mildew dominant resistance gene in the Pm5 locus for high-throughput marker-assisted selection. Plant Dis. 2023;107(2):450–6. [DOI] [PubMed] [Google Scholar]
- 61.Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;172–5.
- 62.Lincoln S, Daly M, Lander E. Constructing genetic maps with MAPMAKER/EXP version 3.0: a tutorial and reference manual. A Whitehead Institute for Biomedical Research Technical Report, Third Edition, Whitehead Institute, Cambridge, MA, USA, 1993.
- 63.Han G, Liu H, Zhu S, Gu T, Cao L, Yan H, Jin Y, Wang J, Liu S, Zhou Y, Shi Z, He H, An D. Two functional CC-NBS-LRR proteins from rye chromosome 6RS confer differential age-related powdery mildew resistance to wheat. Plant Biotechnol J. 2024;22(1):66–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: Additional file 1: Table S1: Primers developed in this study. Table S2: Primers used in this study. Table S3: cDNA sequences of PmDR88_V1 and PmDR88_V2. Table S4: Responses of DR88 and 44 wheat accessions with known powdery mildew resistance genes to 22 different Blumeria graminis f. sp. tritici isolates. Table S5: Detection results of molecular markers JS717xJS718 and WGRE77410 for PmDR88 in 24 cultivars or breeding lines from China.
Supplementary Material 2: Additional file 2: Fig. S1: PCR amplification patterns of the molecular marker JS717xJS718 in 105 F2:3 families from the cross of DR88 and Mo75. (a) M, Trans2K Plus II. 1-84, 25 homozygous resistant and 59 segregating families after inoculating with the Blumeria graminis f. sp. tritici isolate E09. 85, Mo75. 86, DR88. (b) M, Trans2K Plus II. 1-23, 21 homozygous susceptible families after inoculating with the Bgt isolate E09. 22, Mo75. 23, DR88.
Supplementary Material 3: Fig. S2: PCR amplification patterns of molecular markers JS717xJS718 and WGRE77410 in various wheat cultivars/lines from China. M, pBR322/Msp I for JS717xJS718 and Trans2K Plus II for WGRE77410. 1, DR88. 2, Sinxin 6171. 3, Jimai 330. 4, Jimai 340. 5, Henong 826. 6, Hanmai 4. 7, Yumai 35. 8, Yumai 51. 9, Fanyumai 18. 10, Fanyumai 20. 11, Xinong 9718. 12, Zhongmai 5215. 13, Lunxuan 49. 14, Lunxuan 69.
Data Availability Statement
All the data generated or analyzed during the current study were included in the manuscript and its additional files. Sequence data were deposited at NCBI GenBank under the accession numbers PQ550686 (PmDR88_V1 CDS) and PQ550687 (PmDR88_V2 CDS). The raw data and the Bgt isolates used in this study are available from the corresponding author on reasonable request.