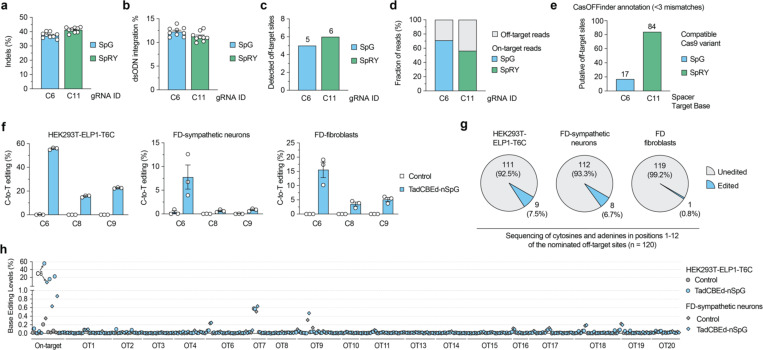
Figure 3. Base editing specificity in correcting the ELP1 T6C mutation.
a-b, Results from GUIDE-seq2 experiments including analysis of insertion or deletion (indels; panel a) and quantification of reads containing the GUIDE-seq2 dsODN tag (panel b) in ELP1 T6C HEK 293T cells when using SpCas9 variants SpG nuclease with gRNA-C6 or SpRY nuclease with gRNA-C11. c, Total number of GUIDE-seq2-detected off-target sites using SpG or SpRY. d, Percentage of total GUIDE-seq reads attributable to the on-target site or cumulative off-target sites. e, Number of putative off-target sites in the human genome with up to 3 mismatches for the spacers of gRNAs C6 and C11, as annotated by CasOFFinder. f, On-target C-to-T editing levels in HEK293T-ELP1-T6C cells, iPSC-derived FD-sympathetic neurons, and FD patient primary fibroblast cells when treated with TadCBEd-nSpG paired with gRNA-C6. g, Pie chart showing a summary of off-target sites validated for gRNA C6 and SpG across all 3 cell types in panel f, based on in silico CasOFFinder and GUIDE-seq2 nominations. h, Summary of on- and off-target base editing levels in three cell lines from panel f. Cells were untreated (naïve) or treated with TadCBEd-nPSG and gRNA C6. Genomic DNA was subjected to PCR for the on-target site and 19 off-target sites (nominated by CasOFFinder and GUIDE-seq2) with data analysis via CRISPResso2 for n = 3 independent biological replicates. For panels a-b and f, mean, standard errors of the mean (s.e.m.), and individual data points are shown for n = 3 or 9 independent biological replicates.