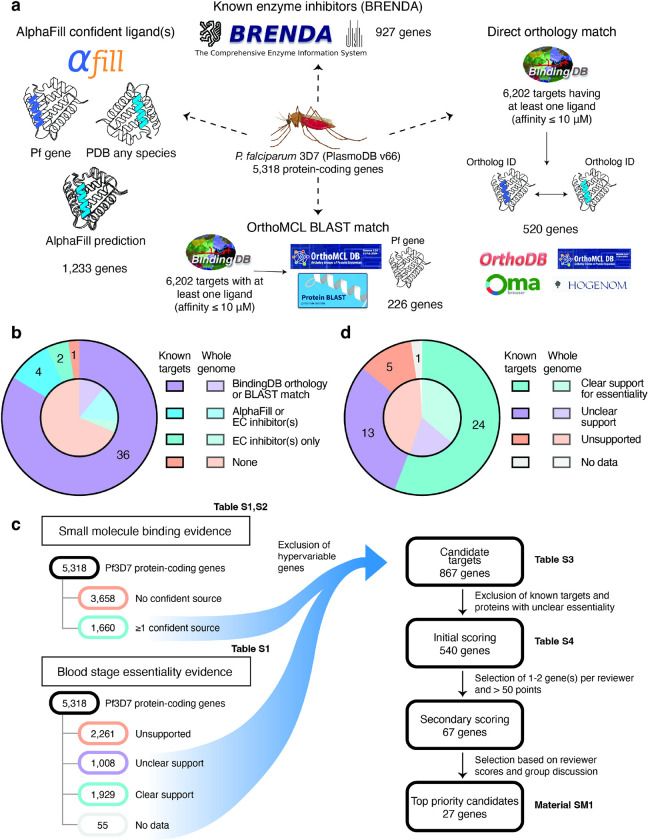
Figure 1. Identification of P. falciparum protein-coding genes that are potentially druggable (n = 1,660) and genes that have evidence of blood stage essentiality (n = 2,992).
a) Identification of potentially druggable genes. Diagram illustrating the four methods used to identify genes with evidence of small molecule binding. Out of the 5,318 protein-coding 3D7 falciparum genes, 226 were found through a BLAST search using BindingDB 6,202 validated drug-targets, and 520 falciparum genes were found through orthology queries with OrthoMCL. A total of 1,233 genes had a confident AlphaFill hit transplant, and 927 falciparum genes were mapped through a validated EC number from BRENDA database. b) Binding evidence for validated targets vs. all genes. Distribution of binding evidence for 43 known targets1 (outer pie) compared to the distribution across all 5,275 P. falciparum 3D7 genes (inner pie). c) Workflow for the identification of 867 candidate targets and subsequent prioritization. Identification and filtering process to define the list of 867 P. falciparum candidate targets. The intersection of 1,660 genes with evidence of small molecule binding and 2,992 genes with essentiality support was used to determine the list of 867 candidates, after excluding hypervariable regions. Subsequent candidate prioritization resulted in 540 candidates that were subjected to initial scoring, 67 of which underwent a second round of scoring, and 27 top ranking targets discussed by a panel of experts that are likely to progress in the near future. d) Essentiality classifications for validated targets vs. all genes. Distribution of essentiality classifications for 43 known targets1 (outer pie) compared to the distribution of 5,275 3D7 genes (inner pie). Essentiality classifications are described in Methods.