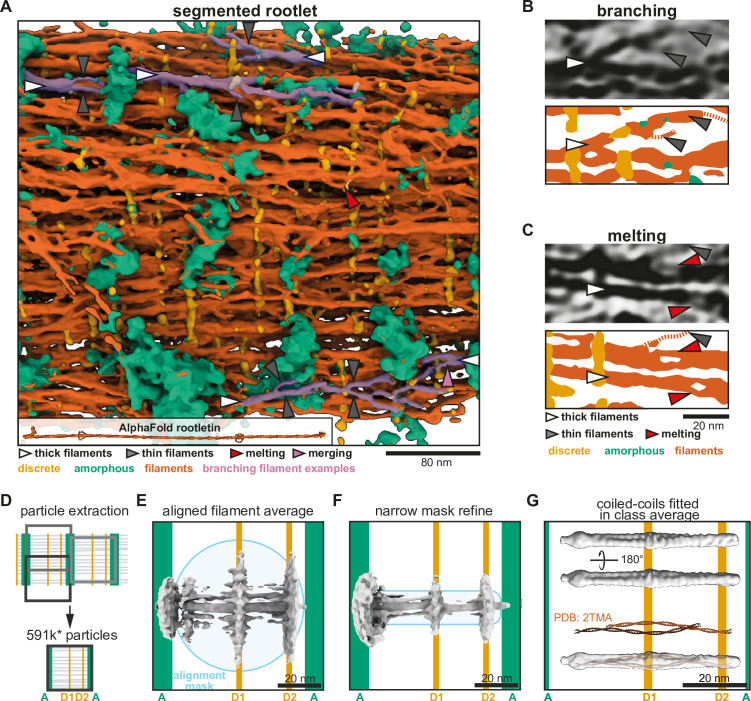
Figure 3. Rootlet filaments are highly flexible and occur as coiled-coil dimers.
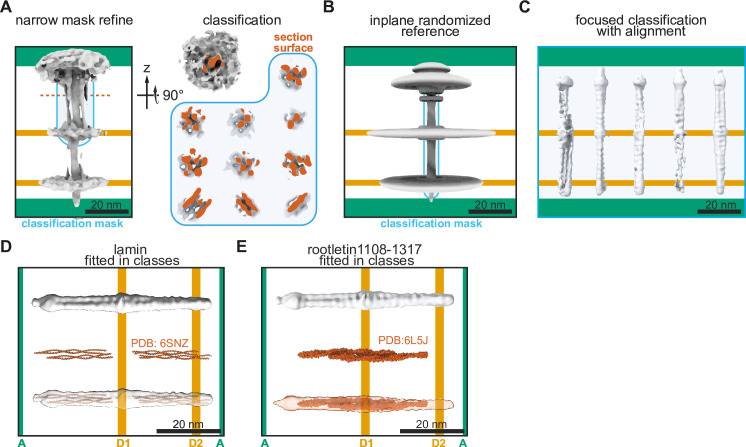
(A) Semi-automated segmentation of a rootlet tomogram. The inset shows a 265-nm long model of stitched rootletin AlphaFold predictions. Filaments that show splaying and merging were manually highlighted in pink. (B, C) Tomogram slices and their corresponding segmentations. (B) Example of thick filaments splaying into thin filaments, each indicated by arrowheads as in the legend of panel C. (C) Example of filament melting pointed out by red arrowheads. (D) Schematic of the location along the rootlet where particles were extracted. (E) The initial average after alignment of 180,252 particles with a wide spherical alignment mask. (F) The initial average of particles aligned with a narrower cylindrical mask. (G) A class average of 34,490 particles, aligned and classified with a narrow mask. The PDB structure (PDB: 2TMA) of two lamin tetramers is shown in red and fitted in the class average.