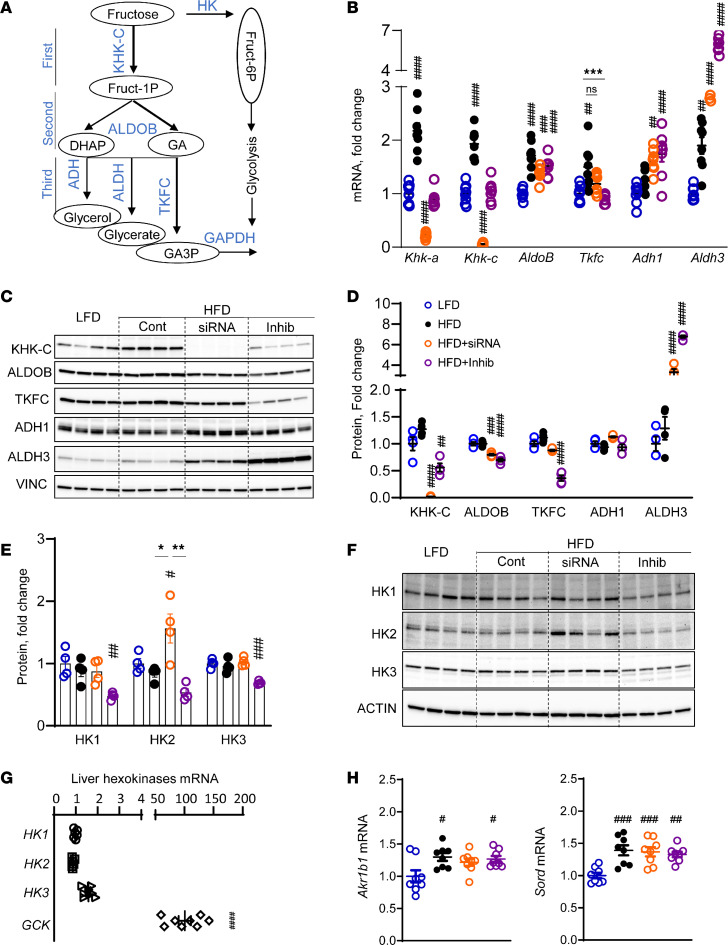
Figure 3. KHK siRNA completely deletes KHK-C and increases HK2, while the inhibitor partially decreases both KHK-C and TKFC proteins.
(A) The fructose metabolism pathway. HK, hexokinase; ALDOB, aldolase B; DHAP, dihydroxyacetone phosphate; GA, glyceraldehyde; ADH, alcohol dehydrogenase; ALDH, aldehyde dehydrogenase; GA3P, glyceraldehyde 3-phosphate. (B) mRNA of fructose-metabolizing enzymes from livers of the mice. n = 6 mice per group. Tkfc, triokinase and FMN cyclase. (C) Western blot and (D) densitometry quantification of fructose metabolizing enzymes in liver lysates. n = 4 mice per group. (E) Densitometry quantification and (F) Western blot of HKs. n = 4 mice per group. (G) mRNA expression of HKs in the liver from mice fed an LFD. (H) Hepatic mRNA expression of aldo-keto reductase (Akr1b1) and sorbitol dehydrogenase (Sord). Statistical analysis was performed using 1-way ANOVA compared with the LFD group (#P < 0.05; ##P < 0.01; ###P < 0.001; ####P < 0.0001) with post hoc 2-tailed t tests between the individual groups (*P < 0.05; **P < 0.01; ***P < 0.001).