Abstract
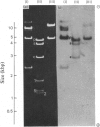
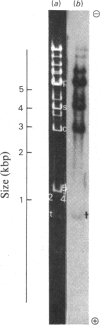
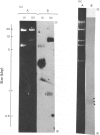
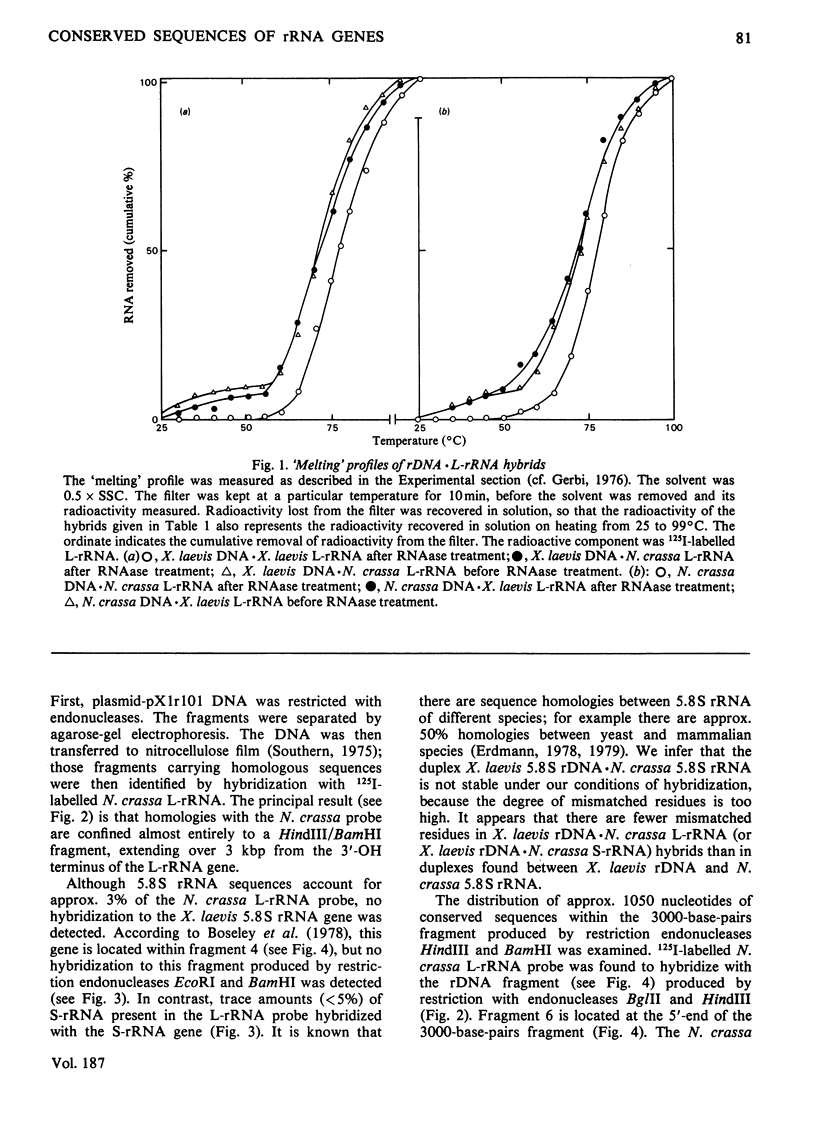
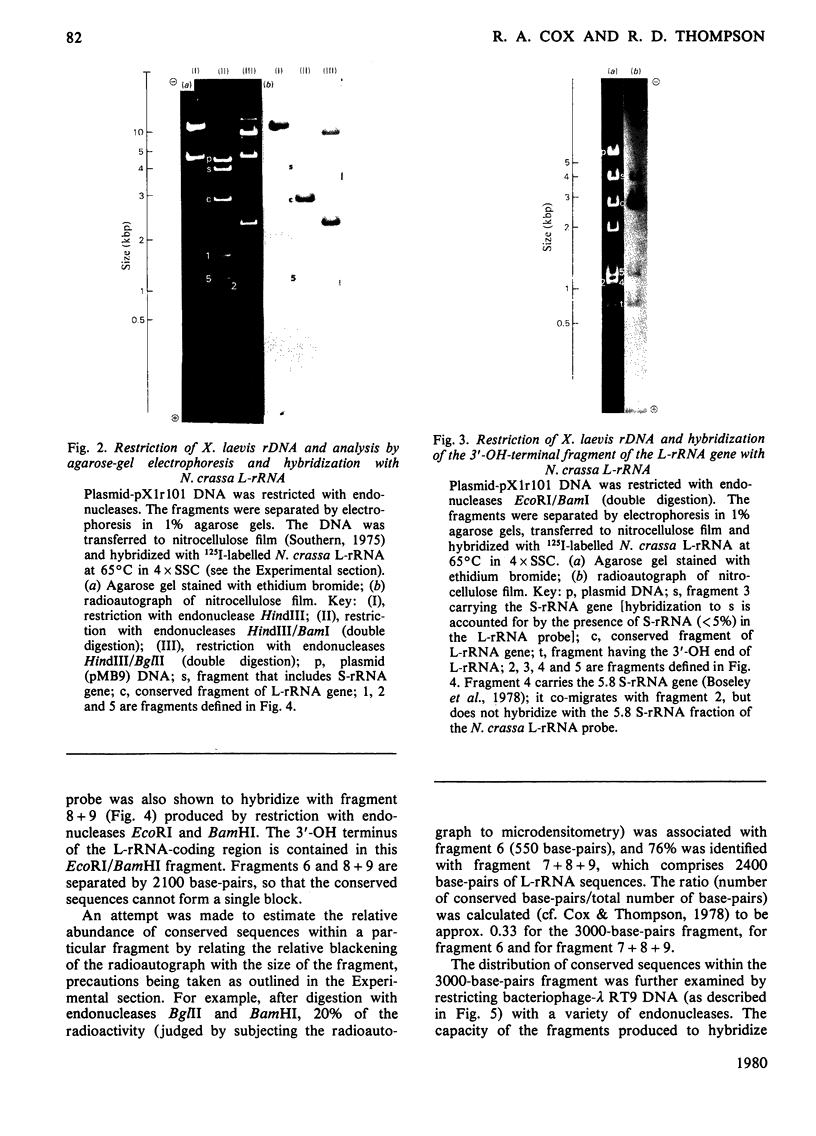
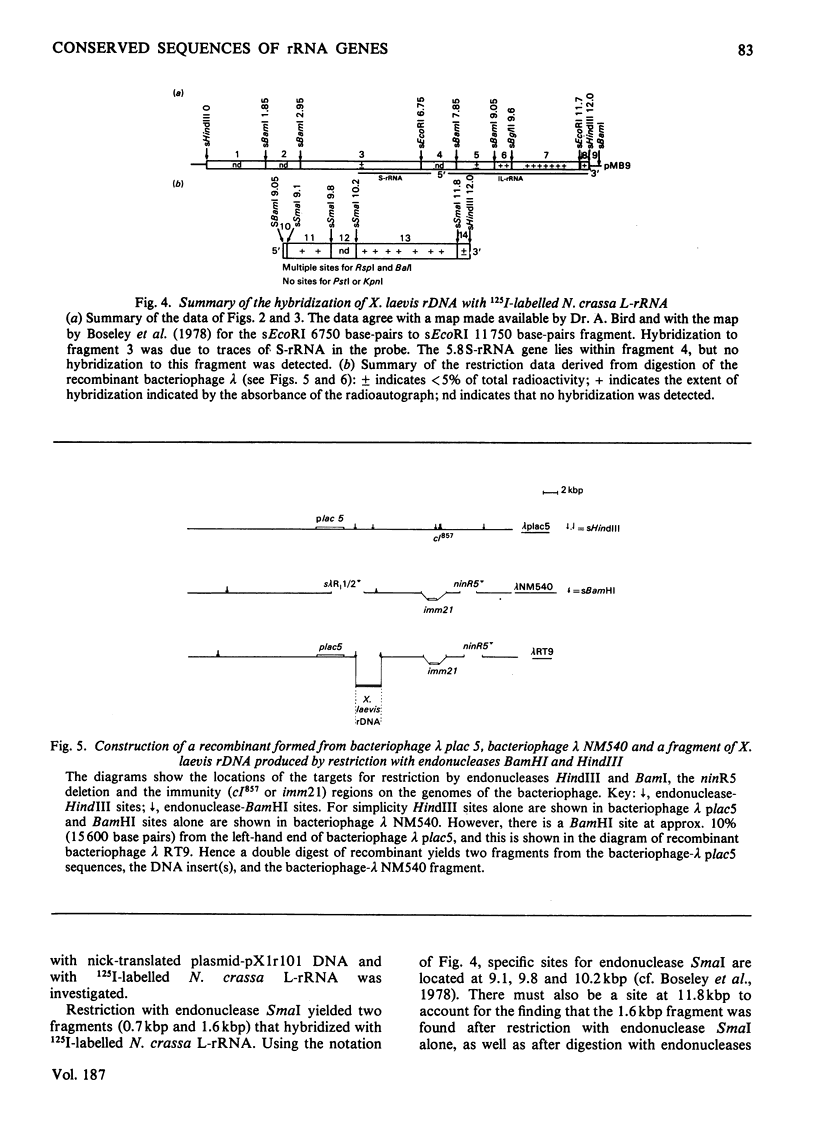
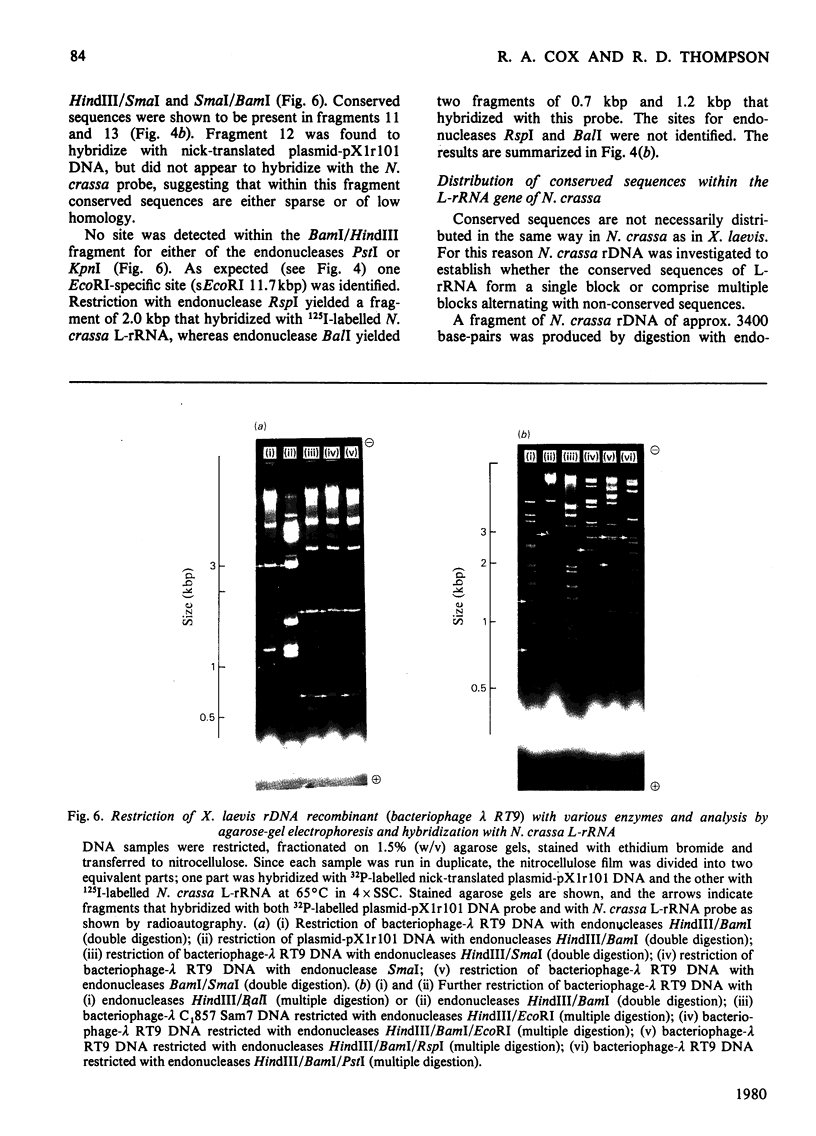
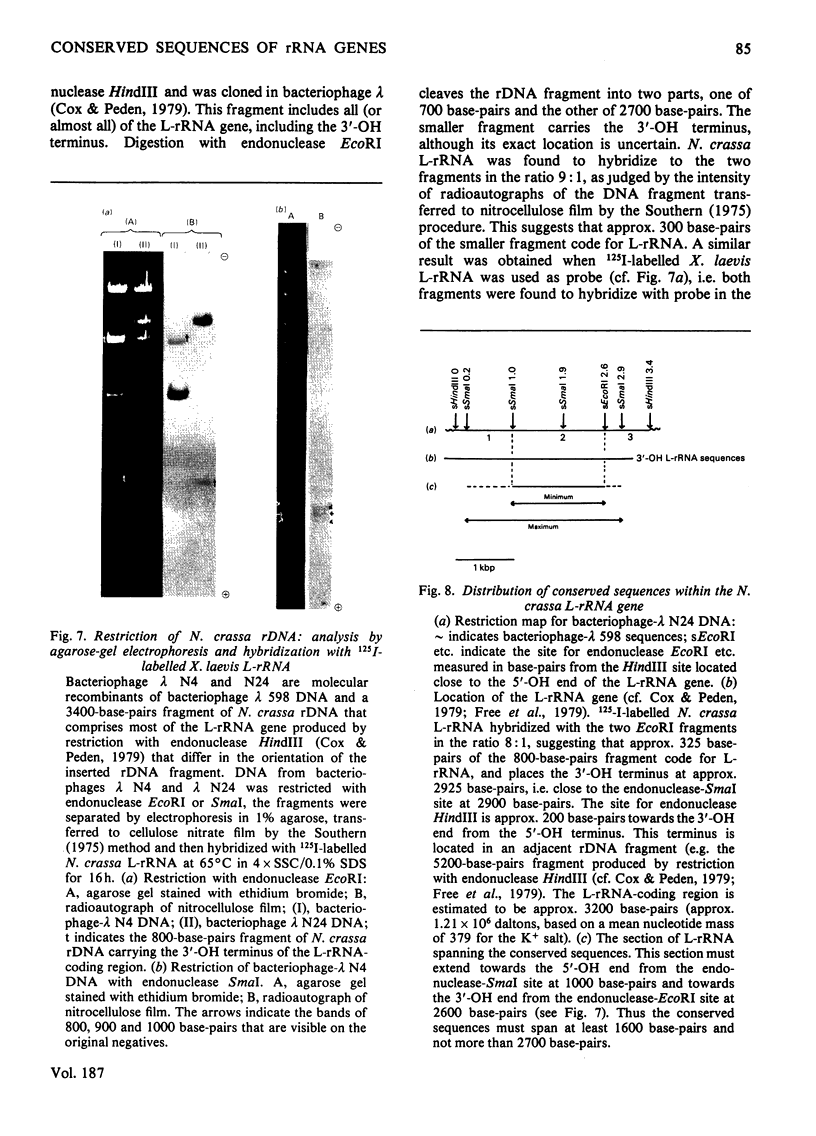
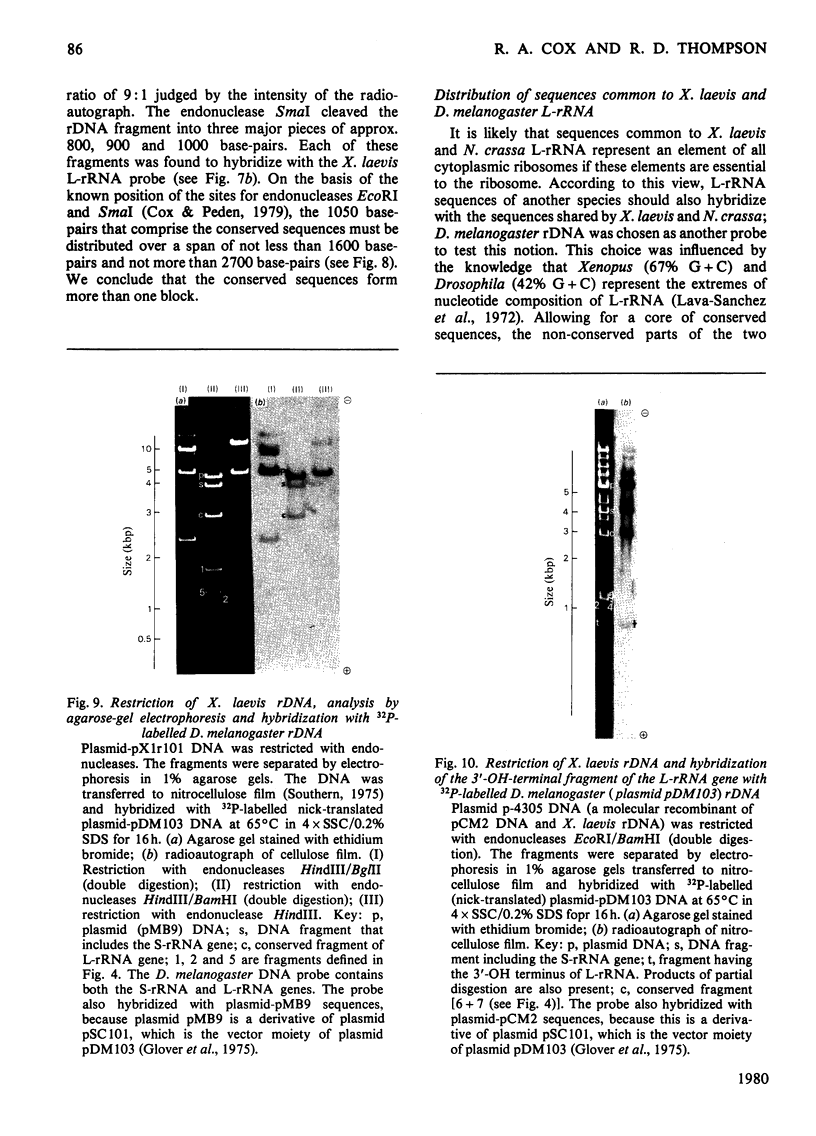
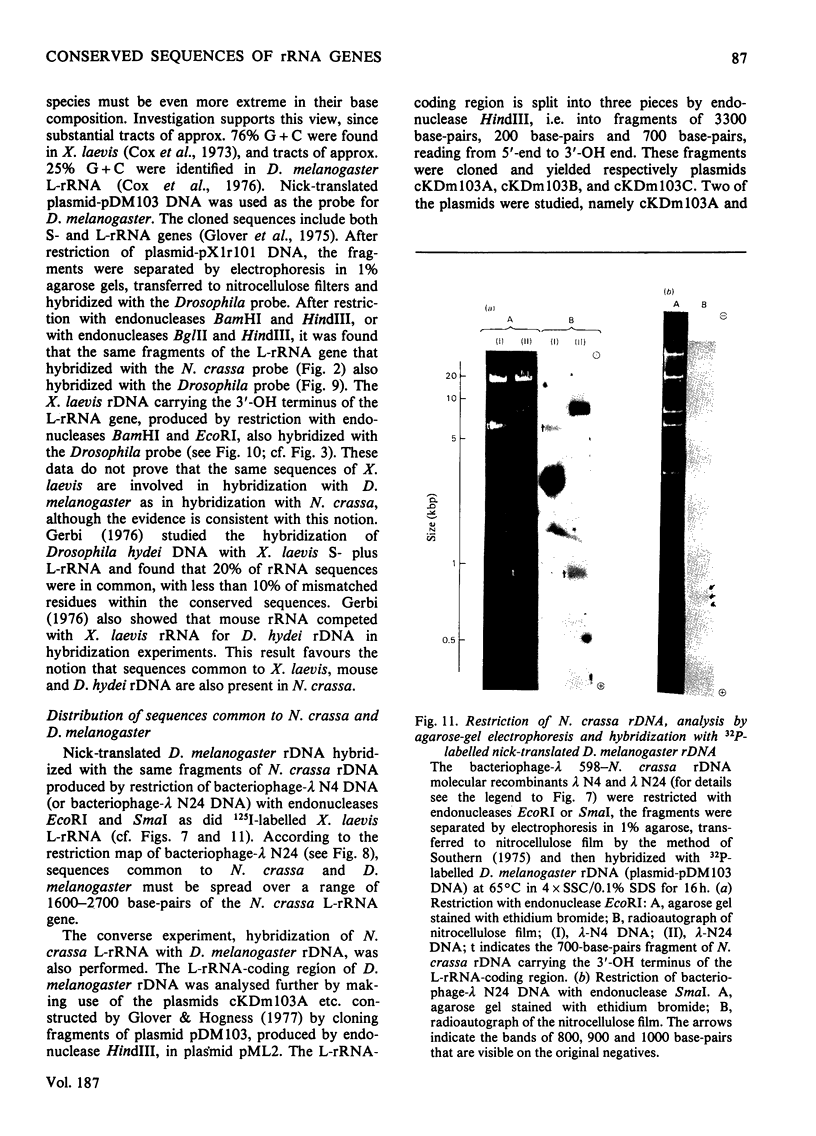
The extent of homology between the nucleotide sequence of L-rRNA (the major RNA component of the larger ribosomal subparticle) of a lower eukaryote (Neurospora crassa) and an amphibian (Xenopus laevis) was investigated by utilizing rDNA (DNA coding for rRNA) of X. laevis cloned in plasmids pMB9 and pML2, and rDNA of N. crassa cloned in bacteriophage lambda. Hybridization studies revealed that sequences common to both N. crassa and X. laevis L-rRNA comprise a total of approx. 1050 /+- 200 nucleotides. The thermal stability of the X. laevis rDNA.N. crassa L-rRNA hybrid was 5 degrees C lower than that of the X. laevis rDNA.X. laevis L-rRNA duplex, indicating the presence of fewer than 10% mismatches in homologous sequences. X. laevis rDNA was analysed by means of restriction endonucleases and hybridization with 125I-labelled N. crassa L-rRNA. Most (at least 95%) of the conserved sequences were present in a 3000-base-pair fragment produced by restriction with endonucleases HindIII and BamHI. This fragment, which includes the 3'-OH terminus of the L-rRNA-coding region, was used as an adaptor in the construction of a bacteriophage-lambda recombinant. One section of the recombinant phage terminating in a HindIII-specific site was obtained from bacteriophage lambda plac5 (after restriction with endonuclease HindIII). A second section terminating in a BamHi-specific site was obtained from bacteriophage lambda 540 (after restriction with endonuclease BamHI). These two parts were joined by means of the X. laevis rDNA fragment. Further analysis of cloned rDNA by means of restriction endonucleases confirmed that conserved sequences were widely distributed throughout the 3000-base-pair fragment produced by HindIII and BamHi endonucleases. A 3400-base-pair fragment of N. crassa rDNA cloned in a bacteriophage lambda [Cox & Peden (1979) Mol. Gen. Genet. 174, 17--24] was restricted with endonucleases. The products were hybridized with 125I-labelled X. laevis L-rRNA. Conserved sequences were shown to be distributed over a range of approx. 1600--2700 base-pairs. Hence, in neither X. laevis nor N. crassa L-rRNA can be conserved sequences from a single block; instead regions of high and low (or no) homology must be intermingled. Both N. crassa rDNA and X. laevis rDNA were found to hybridize with Drosophila melanogaster L-rDNA sequences. Those rDNA fragments with sequences common to X. laevis and N. crassa L-rRNA also hybridized with D. melanogaster L-rRNA probe. Thus the same set of conserved sequences may be present in all three species.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Bukhari A. I. Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases. J Mol Biol. 1975 Mar 15;92(4):529–540. doi: 10.1016/0022-2836(75)90307-1. [DOI] [PubMed] [Google Scholar]
- Bonner T. I., Brenner D. J., Neufeld B. R., Britten R. J. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol. 1973 Dec 5;81(2):123–135. doi: 10.1016/0022-2836(73)90184-8. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitmeyer J. B., Noller H. F. Affinity labeling of specific regions of 23 S RNA by reaction of N-bromoacetyl-phenylalanyl-transfer RNA with Escherichia coli ribosomes. J Mol Biol. 1976 Mar 5;101(3):297–306. doi: 10.1016/0022-2836(76)90149-2. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A., Godwin E., Hastings J. R. Spectroscopic evidence for the uneven distribution of adenine and uracil residues in ribosomal ribonucleic acid of Drosophila melanogaster and of Plasmodium knowlesi and its possible evolutionary significance. Biochem J. 1976 Jun 1;155(3):465–475. doi: 10.1042/bj1550465a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A., Greenwell P., Hirst W. Re-activation of the peptidyltransferase centre of rabbit reticulocyte ribosomes after inactivation by exposure to low concentrations of magnesium ion. Biochem J. 1976 Dec 15;160(3):521–531. doi: 10.1042/bj1600521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A., Hirst W. A study of the influence of magnesium ions on the conformation of ribosomal ribonucleic acid and on the stability of the larger subribosomal particle of rabbit reticulocytes. Biochem J. 1976 Dec 15;160(3):505–519. doi: 10.1042/bj1600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A., Peden K. A study of the organisation of the ribosomal ribonucleic acid gene cluster of Neurospora crassa by means of restriction endonuclease analysis and cloning in bacteriophage lambda. Mol Gen Genet. 1979 Jul 2;174(1):17–24. doi: 10.1007/BF00433300. [DOI] [PubMed] [Google Scholar]
- Cox R. A., Peden K. Organization of the ribosomal ribonucleic acid gene cluster of Neurospora crassa by means of restriction endonucleases and cloning in bacteriophage lambda [proceedings]. Biochem Soc Trans. 1978;6(6):1230–1232. doi: 10.1042/bst0061230. [DOI] [PubMed] [Google Scholar]
- Cox R. A. Structure and function of prokaryotic and eukaryotic ribosomes. Prog Biophys Mol Biol. 1977;32(3):193–231. [PubMed] [Google Scholar]
- Cox R. A., Thompson R. A novel recombinant of phage lambda and a conserved 3000-base-pair fragment of Xenopus laevis ribosomal deoxyribonucleic acid produced by restriction with endonucleases Hind III/BamI [proceedings]. Biochem Soc Trans. 1978;6(6):1232–1233. doi: 10.1042/bst0061232. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K. A reinvestigation of 5' leads to 3' polarity in 40S ribosomal RNA precursor of Xenopus laevis. Cell. 1976 Jul;8(3):443–448. doi: 10.1016/0092-8674(76)90157-4. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Collection of published 5S and 5.8S RNA sequences and their precursors. Nucleic Acids Res. 1979 Jan;6(1):r29–r44. doi: 10.1093/nar/6.1.419-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free S. J., Rice P. W., Metzenberg R. L. Arrangement of the genes coding for ribosomal ribonucleic acids in Neurospora crassa. J Bacteriol. 1979 Mar;137(3):1219–1226. doi: 10.1128/jb.137.3.1219-1226.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi S. A. Fine structure of ribosomal RNA. I. Conservation of homologous regions within ribosomal RNA of eukaryotes. J Mol Biol. 1976 Sep 25;106(3):791–816. doi: 10.1016/0022-2836(76)90265-5. [DOI] [PubMed] [Google Scholar]
- Gillespie D., Spiegelman S. A quantitative assay for DNA-RNA hybrids with DNA immobilized on a membrane. J Mol Biol. 1965 Jul;12(3):829–842. doi: 10.1016/s0022-2836(65)80331-x. [DOI] [PubMed] [Google Scholar]
- Glover D. M., Hogness D. S. A novel arrangement of the 18S and 28S sequences in a repeating unit of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):167–176. doi: 10.1016/0092-8674(77)90212-4. [DOI] [PubMed] [Google Scholar]
- Glover D. M., White R. L., Finnegan D. J., Hogness D. S. Characterization of six cloned DNAs from Drosophila melanogaster, including one that contains the genes for rRNA. Cell. 1975 Jun;5(2):149–157. doi: 10.1016/0092-8674(75)90023-9. [DOI] [PubMed] [Google Scholar]
- Godwin E., Cox R. A., Huvos P. Studies of the RNA and protein moieties of the larger subribosomal particle of rabbit reticulocytes. Acta Biol Med Ger. 1974;33(5-6):733–752. [PubMed] [Google Scholar]
- HORWITZ J. P., CHUA J., CURBY R. J., TOMSON A. J., DAROOGE M. A., FISHER B. E., MAURICIO J., KLUNDT I. SUBSTRATES FOR CYTOCHEMICAL DEMONSTRATION OF ENZYME ACTIVITY. I. SOME SUBSTITUTED 3-INDOLYL-BETA-D-GLYCOPYRANOSIDES. J Med Chem. 1964 Jul;7:574–575. doi: 10.1021/jm00334a044. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Lava-Sanchez P. A., Amaldi F., Posta A. L. Base composition of ribosomal RNA and evolution. J Mol Evol. 1972 Dec 29;2(1):44–55. doi: 10.1007/BF01653942. [DOI] [PubMed] [Google Scholar]
- Loening U. E. Molecular weights of ribosomal RNA in relation to evolution. J Mol Biol. 1968 Dec;38(3):355–365. doi: 10.1016/0022-2836(68)90391-4. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T. E., Bicknell J. N., Kumar A. Hybrid 80S monomers formed from subunits of ribosomes from protozoa, ungi, plants, and mammals. Biochem Genet. 1970 Oct;4(5):603–615. doi: 10.1007/BF00486098. [DOI] [PubMed] [Google Scholar]
- Murray K., Murray N. E. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease III of Haemophilus influenzae and restriction endonuclease I of Escherichia coli. J Mol Biol. 1975 Nov 5;98(3):551–564. doi: 10.1016/s0022-2836(75)80086-6. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Murray K. Manipulation of restriction targets in phage lambda to form receptor chromosomes for DNA fragments. Nature. 1974 Oct 11;251(5475):476–481. doi: 10.1038/251476a0. [DOI] [PubMed] [Google Scholar]
- Pratt H., Cox R. A. Dissociation of ribosomes from oocytes of Xenopus laevis into active subparticles. Biochem J. 1971 Oct;124(5):897–903. doi: 10.1042/bj1240897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J. H., Brown D. D. Retention of common nucleotide sequences in the ribosomal deoxyribonucleic acid of eukaryotes and some of their physical characteristics. Biochemistry. 1971 Jul 6;10(14):2761–2769. doi: 10.1021/bi00790a017. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]