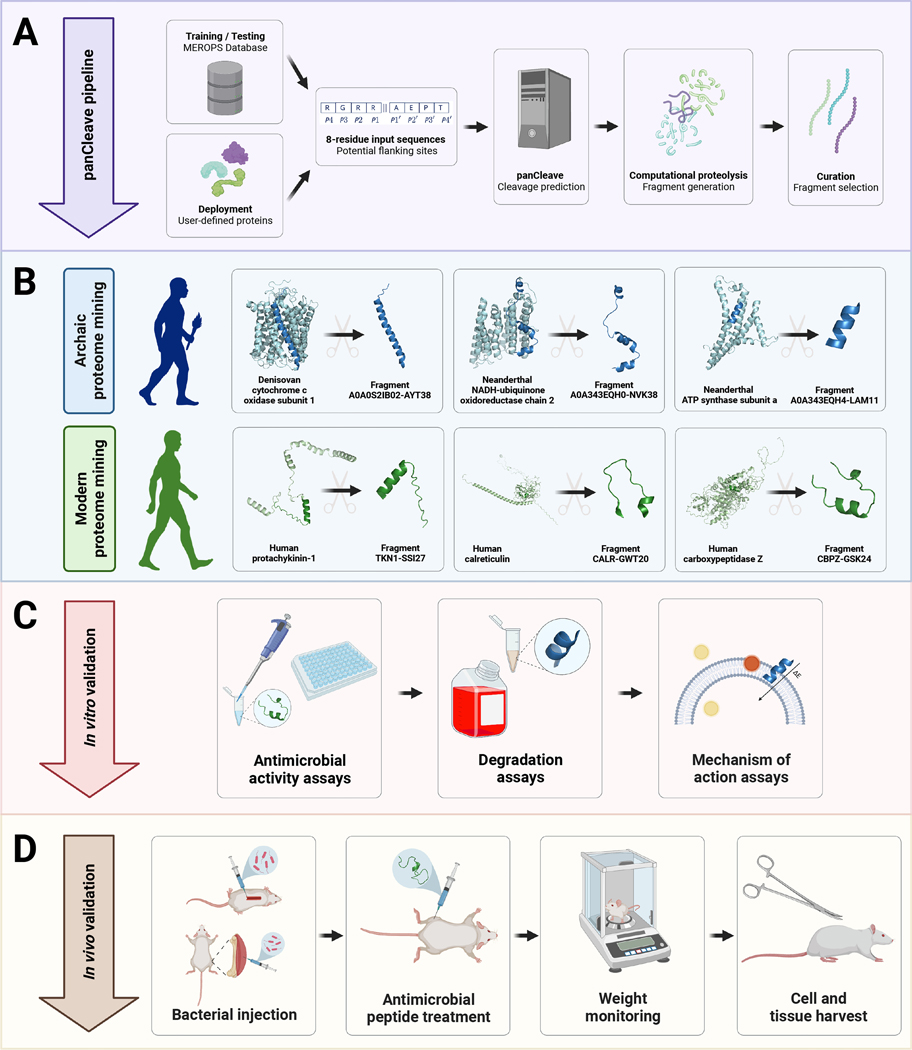
Fig. 1. Computational-experimental framework for molecular de-extinction of antimicrobial peptides.
Panel (A) demonstrates the computational proteolysis pipeline, where user-defined proteins are processed into 8-residue subsequences that are classified as cleavage and non-cleavage sites. Input proteins are then tokenized at predicted cleavage sites, and the resulting fragments can be filtered by user-defined curation methods. Curation methods can include machine learning-based activity prediction, human expert curation, or other methods. Successes in archaic and modern proteome mining are visualized in panel (B), where precursors were computationally digested to reveal encrypted antimicrobial subsequences. The pipeline concludes with in vitro (C) and in vivo (D) experimental validation of fragment bioactivity, including proteolytic degradation assays, MoA assays, and mouse weight monitoring as a proxy for host toxicity. Figure created with BioRender.com and the PyMOL Molecular Graphics System, Version 2.1 Schrödinger, LLC.