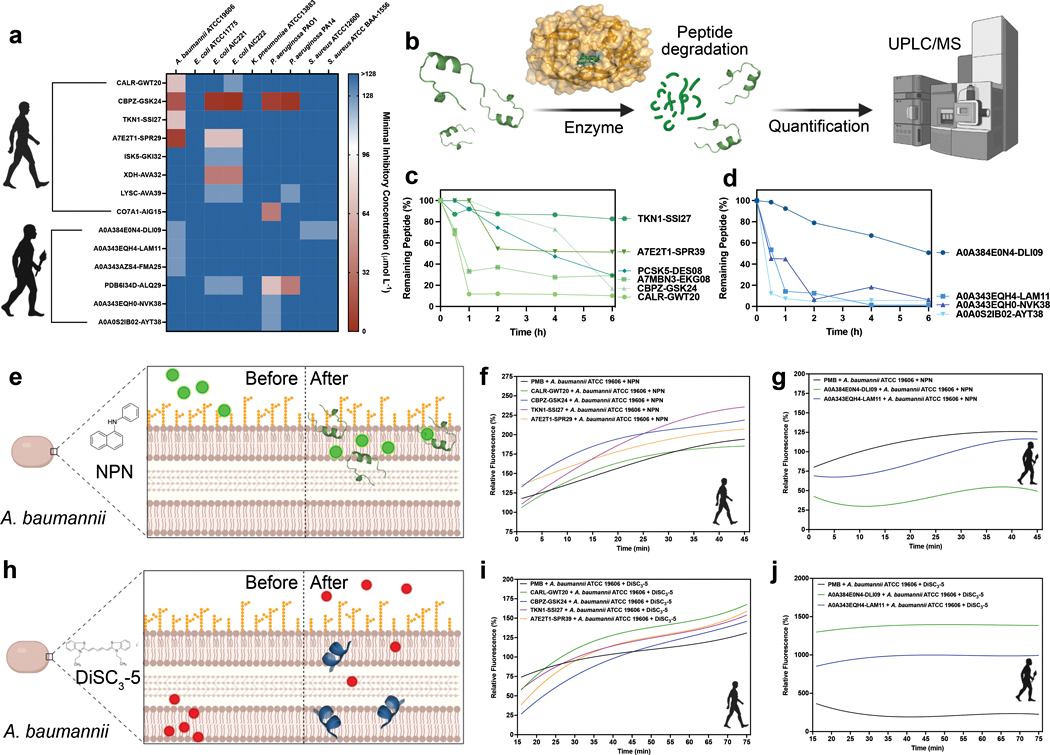
Fig. 3. Antimicrobial activity, resistance to enzymatic degradation, and mechanism of action of modern and archaic EPs.
(a) Antimicrobial activity of the EPs. Briefly, a fix number of 106 bacterial cells per mL−1 was used in all the experiments. The modern and archaic EPs were two-fold serially diluted ranging from 128 to 2 μmol L−1 in a 96-wells plate and incubated at 37 °C for one day. After the exposure period, the absorbance of each well was measured at 600 nm. Untreated solutions were used as controls and minimal concentration values for complete inhibition were presented as a heat map of antimicrobial activities (μmol L−1) against nine pathogenic bacterial strains. All the assays were performed in three independent replicates and the heat map shows the mode obtained within the two-fold dilutions concentration range studied. (b) Schematic of the resistance to enzymatic degradation experiment, where peptides were exposed for a total period of six hours to fetal bovine serum that contains several active proteases. Aliquots of the resulting solution were analyzed by liquid chromatography coupled to mass spectrometry. (c) Modern and (d) archaic peptides had different degradation behaviors. In summary, archaic peptides are more resistant to enzymatic degradation than modern peptides. Experiments were performed in two independent replicates. (e) Schematic showing the behavior of 1-(N-phenylamino)naphthalene (NPN) the fluorescent probe used to indicate membrane permeabilization caused by the EPs. (f) Modern and (g) archaic EPs fluorescence values relative to the untreated control showing that MEPs are more efficient at permeabilizing the outer membrane of A. baumannii cells than polymyxin B (PMB) and archaic EPs. (h) Schematic of how 3,3′-dipropylthiadicarbocyanine iodide [DiSC3-(5)], a hydrophobic fluorescent probe, was used to indicate membrane depolarization caused by the EPs. (i) Modern and (j) archaic EPs fluorescence values relative to the untreated control showing that archaic peptides are much stronger depolarizers of the cytoplasmic membrane of A. baumannii cells than polymyxin B (PMB) and modern EPs. Experiments were performed in three independent replicates. Figure created with BioRender.com and the PyMOL Molecular Graphics System, Version 2.1 Schrödinger, LLC.