Figure 6: Human LUBAC deficiencies cause dysregulation in B cell activation and death.
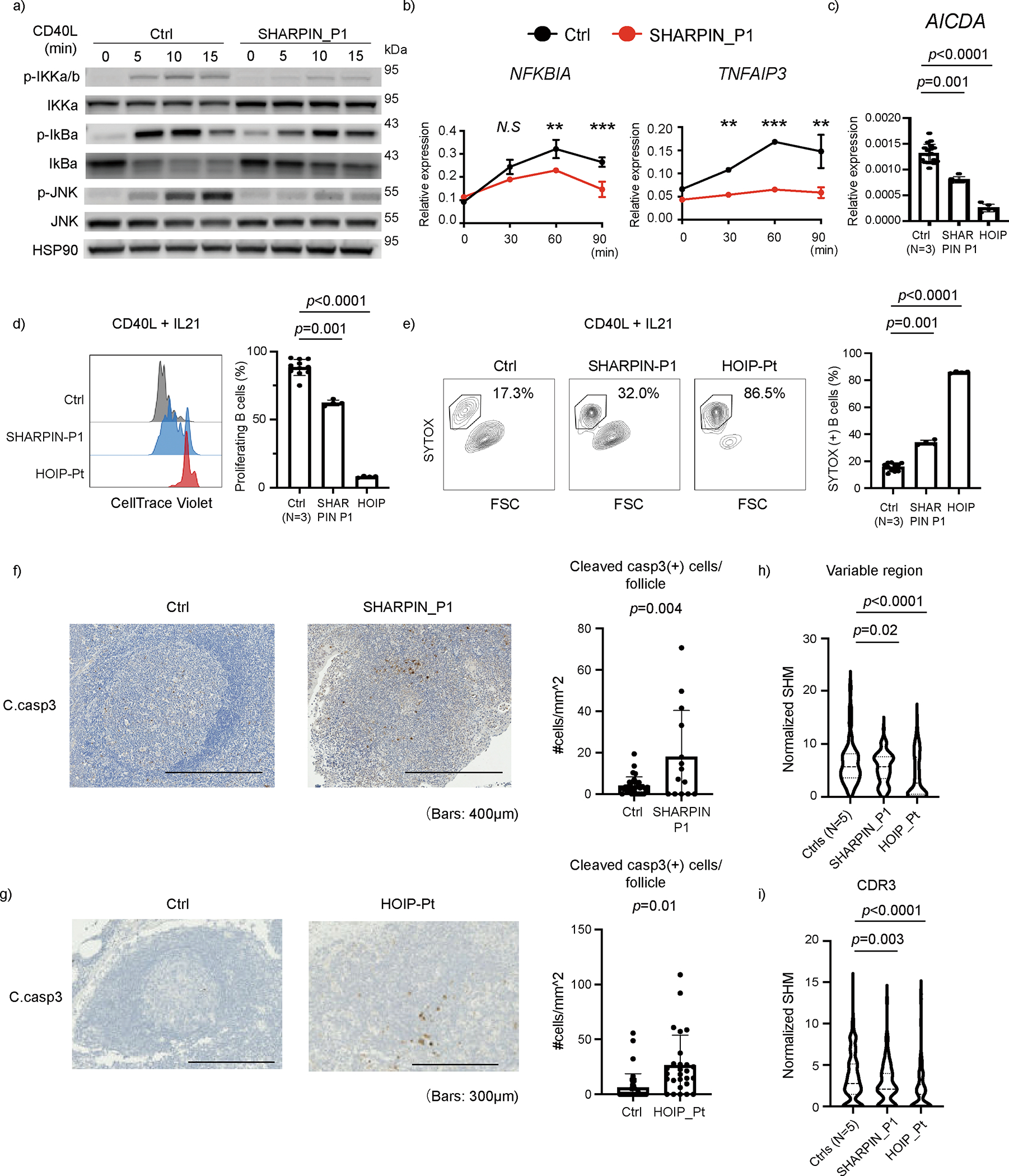
(a) NF-κB induction assay. EBV-immortalized lymphoblast (EBV-B) cells from SHARPIN deficient P1 showed attenuated phosphorylation of IKKα/β, IκBα and JNK after CD40L stimulation. (b) mRNA expression of NF-κB target genes in EBV-B cells after CD40L stimulation. The experiment was performed in biological triplicates, and the expression levels were normalized to GAPDH. Mean values ± s.d are displayed. Significance calculated with two-tailed Student’s t-test. (c) mRNA expression of AICDA gene (encoding AID), normalized to GAPDH. CD19+ primary B cells were enriched by anti-CD19 magnetic beads and stimulated with CD40L for 24h. (d) Proliferation assay using primary B cells from LUBAC deficient patients. PBMCs were stained with CellTrace Violet, cultured with CD40L and IL21 for 96 h and analyzed by flow cytometer. (e) Cell death assay by SYTOX staining of primary B cells from LUBAC deficient patients. PBMCs were cultured with CD40L and IL-21 for 96 h. (c-e) The experiments were performed using biological quadruplicates from a SHARPIN- and a HOIP-deficient patient, and the results were compared with samples from three unrelated healthy donors. Mean values ± s.d are displayed. Significance calculated with one-way ANOVA followed by Tukey-Kramer test. (f-g) Cleaved caspase-3 immunohistochemistry staining of (f) adenoid from SHARPIN deficient patient and (g) axillary lymph node from a HOIP deficient patient, compared with tissues from control donors. The number of cleaved caspase-3 positive cells per follicle was quantified and normalized by follicle area size. Mean values ± s.d are displayed. (h-i) Somatic hypermutation (SHM) quantification in FACS-sorted memory B cells from peripheral blood. SHM in the entire V region (h) and the CDR3 region (i) of the t gene were normalized by the nucleotide length of each clonotype. Normalized SHM of top 100 IGHG clonotypes per each sample are demonstrated. Significance calculated with one-way ANOVA followed by Tukey-Kramer test. (a-e) These ex vivo data are representative of two independent experiments. (f-i) The experiments were not repeated due to the limited clinical specimens.
