Fig. 3. Robust identification of human kidney meQTL.
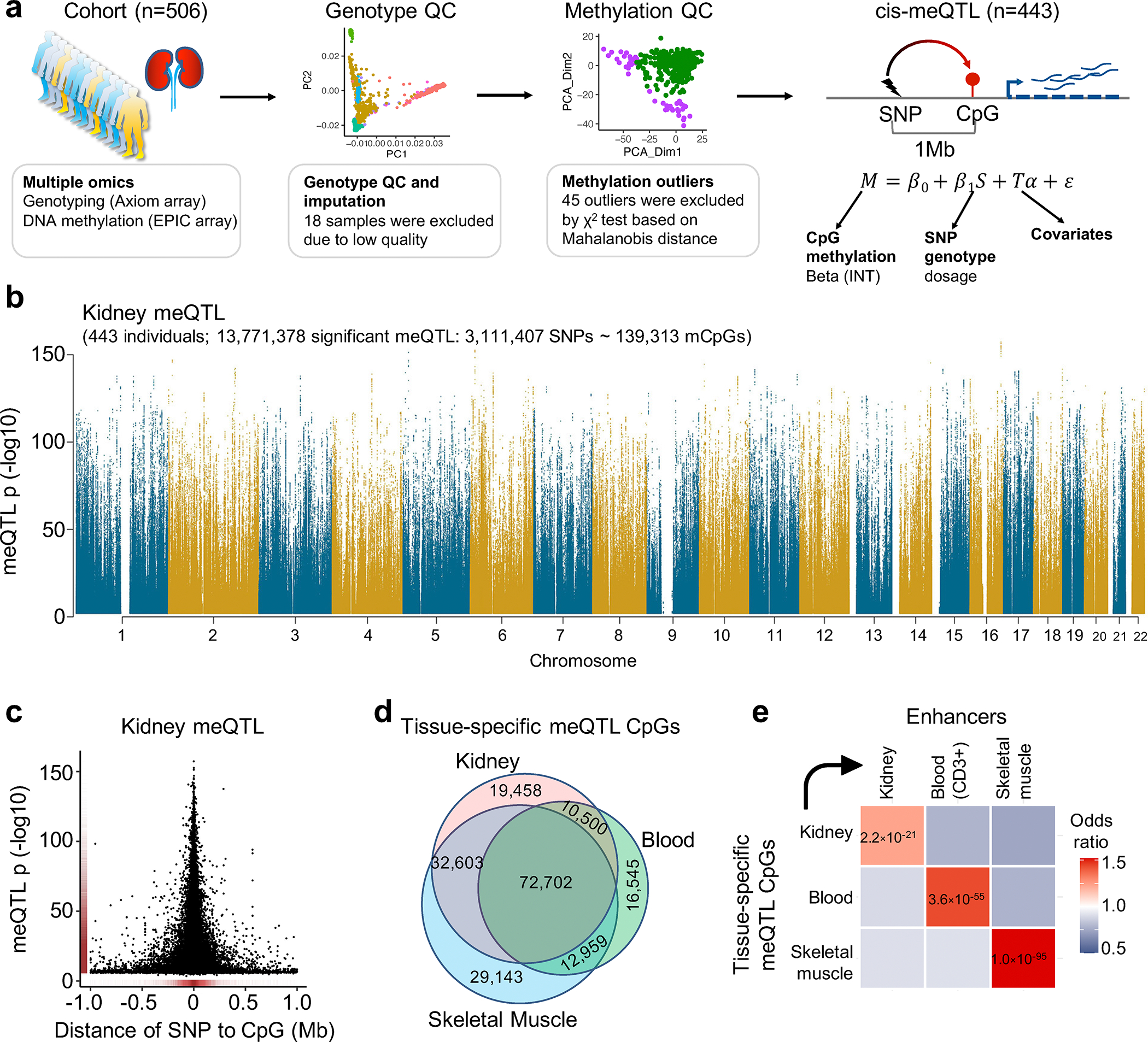
a. Samples and analytical workflow for the meQTL analyses.
b. Manhattan plot of human kidney meQTL data (n=443). X-axis is chromosomal location. Y-axis strength of association -log10(two-sided p). The two-sided p value was calculated by linear regression meQTL model. Only meQTLs with p < 0.01 are shown in the Manhattan plot.
c. The strength of association (-log10(two-sided p), y-axis) of the best mSNPs (the lead meQTL for each mCpG) decreases with the increasing distance (x-axis) from the CpG sites.
d. Overlap of meQTL CpGs between kidney, blood, and skeletal muscle.
e. Enrichment of tissue-specific meQTL CpGs in tissue enhancers. X-axis tissue enhancer, y-axis tissue meQTLs. The color shows the enrichment odds ratio from low (blue) to high (red), while the p-value (two-sided chi-square test) is listed in the box.
