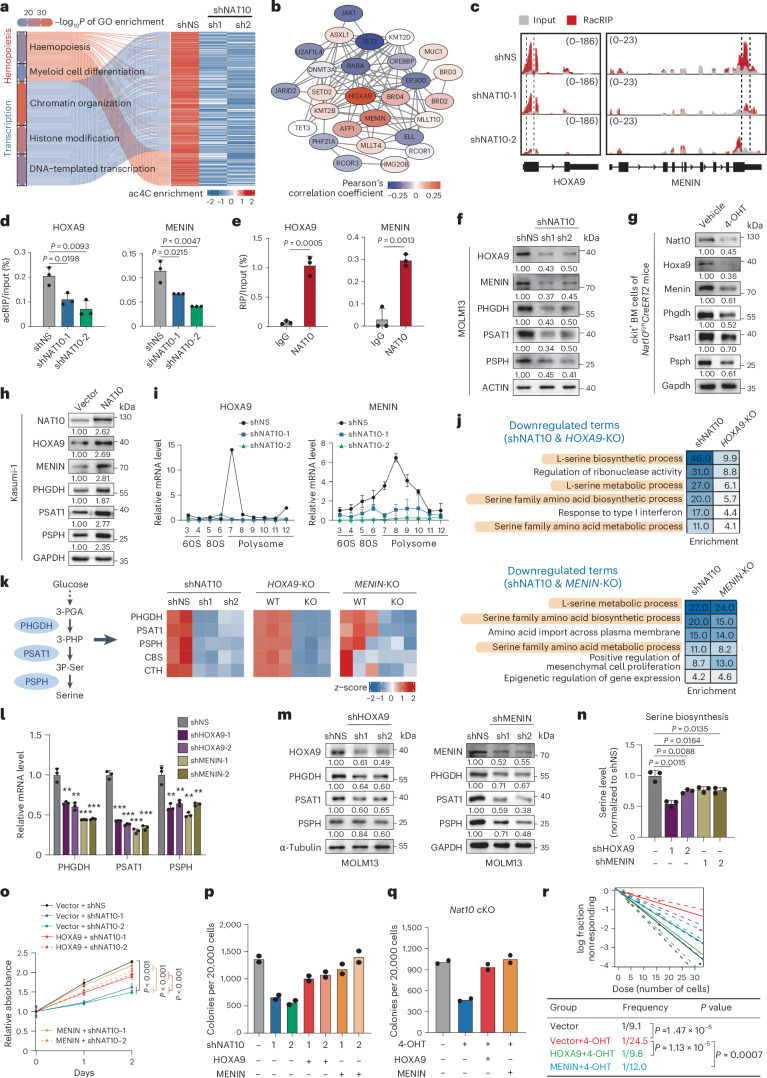
Fig. 5. NAT10 promotes SSP genes transcription and serine biosynthesis through HOXA9/MENIN.
a, Heatmap of hypo-acetylation genes related to transcription regulation and haematopoietic differentiation in NAT10 knockdown MOLM13 cells. b, The interactome of NAT10 targets that are essential for leukaemia transcriptional regulation. The colour of circles represents the expression correlation between NAT10 targets and SSP genes. c, IGV tracks showing ac4C peak distribution in HOXA9 and MENIN mRNAs. High-confidence ac4C sites were marked in dashed area. The y axis represents CPM. d, acRIP–qPCR detecting ac4C levels of HOXA9 and MENIN mRNAs in NAT10 knockdown MOLM13 cells. e, RIP–qPCR assays showing the binding of endogenous NAT10 on HOXA9 and MENIN mRNAs. f–h, Western blotting detecting NAT10 targets in NAT10 knockdown MOLM13 cells (f), in c-Kit+ BM cells from Nat10-cKO mice (g) and in Kasumi-1 stable lines with NAT10 overexpression (h). i, Polysome profiling showing ribosome occupancy on HOXA9 and MENIN mRNAs in MOLM13 cells. j, Commonly enriched GO terms of the downregulated DEGs in NAT10 knockdown and HOXA9-KO/MENIN-KO MOLM13 cells. k, Heatmap showing the expression levels of genes involved in serine metabolism in NAT10 knockdown, HOXA9-KO/MENIN-KO MOLM13 cells. l,m, RT–qPCR (l) and western blotting (m) showing the decrease of mRNA and protein levels of PHGDH, PSAT1 and PSPH in MOLM13 upon HOXA9 or MENIN knockdown. **P < 0.01. ***P < 0.001. n, Quantification of serine biosynthesis levels in MOLM13 upon HOXA9 or MENIN knockdown. o,p, MTT assays (o) and CFA assays (p) in NAT10 knockdown or control MOLM13 cells with or without HOXA9 or MENIN overexpression. Precise P values are shown in Source Data. q,r, CFA (q) and LDA (r) using c-Kit+ BM cells from Nat10-cKO mice transduced with MLL-AF9 plus MSCV-PIG (vector), MSCV-HOXA9 or MSCV-MENIN retroviruses. Values are mean ± s.d. of n = 4 biological replicates in o or n = 3 biological replicates in d, e, i, l and n. Values are mean of n = 2 biological replicates in p and q. Two-tailed Student’s t-tests were used in d, e, l, n and o. A two-sided chi-squared test was used in r.