Abstract
Incompatible plasmids play a crucial role in the horizontal transfer of antibiotic resistance in bacteria, particularly in Gram-negative bacteria, and have thus attracted considerable attention in the field of microbiological research. In the 1970s, these plasmids, housing an array of resistance genes and genetic elements, were predominantly discovered. They exhibit a broad presence in diverse host bacteria, showcasing diversity in geographic distribution and the spectrum of antibiotic resistance genes. The complex genetic structure of plasmids further accelerates the accumulation of resistance genes in Gram-negative bacteria. This article offers a comprehensive review encompassing the discovery process, host distribution, geographic prevalence, carried resistance genes, and the genetic structure of different types incompatible plasmids, including IncA, IncC, IncF, IncL, IncM, IncH, and IncP. It serves as a valuable reference for enhancing our understanding of the role of these different types of plasmids in bacterial evolution and the dissemination of antibiotic resistance.
Keywords: genetic structures, plasmids, horizontal gene transfer, resistance genes, genetic element
1. Introduction
In modern medicine and clinical practice, widely used antimicrobials and medications have significantly improved human health. However, the emergence of antibiotic resistance poses severe challenges to public health and clinical treatment (Di Pilato et al., 2024). Typically, Escherichia coli bacterial genetic material contains numerous resistance genes, which enable the host bacteria to express antibiotic resistance. Plasmids, as extrachromosomal genetic elements, can undergo horizontal transfer among bacterial strains. Therefore, resistance genes carried by plasmids play an essential role in the horizontal transfer of antibiotic resistance genes that cannot be ignored. In 1952, Joshua Lederberg first proposed the concept of plasmids, initiating discussions on the systematic classification of plasmid systems (Lederberg, 1952). García-Bartels et al. provided a comprehensive review of the history of plasmid classification. Major plasmid classification schemes include fi+ and fi- plasmid typing (based on conjugative transfer ability), superinfection immunity typing (based on exclusion phenomena), incompatibility plasmid replicon typing (based on replicons), MOB typing (based on relaxase genes), and classification schemes based on plasmid whole genome analysis (Garcillán-Barcia et al., 2023). In accordance with the PBRT typing method, incompatible plasmids are classified into 27 types (Villa and Carattoli, 2020). Given the widespread distribution and conjugative potential of IncA, IncC, IncF, IncL, IncM, IncH, and IncP plasmids in Gram-negative bacteria, this review offers a comprehensive analysis of their characteristics. Additionally, other types of incompatible plasmids (such as IncN, IncI, IncQ, IncX, etc.) are also crucial in microbial resistance. The presence of these plasmids reflects the diverse genetic strategies that bacteria adapt to the environment and antibiotic pressure. The resistance genes carried by these plasmids are closely related to the origin and ecological environment of the strains, playing crucial roles in the transmission and evolution of resistance (Tobin et al., 2024). However, as plasmids spread, the carried resistance genes also show a gradual evolution. Under different time and environmental conditions, plasmids can capture new resistance genes, and they may also lose existing ones (Ghaly et al., 2022). This evolution not only reflects the continuous adaptation of bacteria to antibiotic pressure but also has profound implications for the evolution of resistance genes. Therefore, we need in-depth research on the evolutionary dynamics of these resistance genes to better understand the formation and development of bacterial resistance to antimicrobials.
2. The primary mechanisms of antibiotic resistance in bacteria
The detection rates of multidrug-resistant (MDR), extensively drug-resistant (XDR), and pandrug-resistant (PDR) strains are continuously increasing, closely associated with the overuse of antibiotics and the spread of resistance genes mediated by mobile genetic elements. Bacteria express antimicrobials resistance primarily through three major mechanisms. Firstly, they reduce the intracellular concentration of antibiotics. For example, Gram-negative bacteria decrease antibiotic permeability by producing biofilms and actively expel antimicrobials via efflux pumps such as OqxAB and TmexCD-ToprJ.Secondly, they alter antibiotic targets. Recent studies have found that mutations in the KPC-2 resistance gene, leading to KPC-33 or KPC-189, confer resistance to ceftazidime and avibactam (Zhang et al., 2024). Lastly, bacteria inactivate antimicrobials by producing hydrolytic enzymes (Blair et al., 2015). β-lactam antibiotics are widely used for infections caused by Gram-negative Enterobacteriaceae, thus resistance problems arising from β-lactamases produced by bacteria have garnered significant attention. β-lactamases are mainly classified into four classes: A, B, C, and D. Among them, serine β-lactamases are classified into three types: Class A (e.g., KPC, SHV, TEM, and CTX-M), Class C (e.g., AmpC), and Class D (e.g., OXA), while Class B refers to metallo-β-lactamases, with common examples including VIM, IMP, and NDM (Karampatakis et al., 2023). Furthermore, there is a growing trend in the resistance to aminoglycosides, phosphomycin, and polymyxins (Luo et al., 2024). The resistance mechanism of aminoglycosides is primarily associated with bacterial production of 16S rRNA methyltransferases and is often clinically managed by combination therapy with β-lactam antibiotics to control infections (Yang and Hu, 2022). The resistance mechanism of phosphomycin is mainly related to defects in transport proteins, modifications of targets, and inactivation mediated by fosA ( Huang et al., 2021). Polymyxins are often considered the last line of defense against infections; however, the increasing resistance rate is a cause for significant concern. Its resistance is associated with multiple genes, including chromosomal genes pmrCAB, phoPQ, mgrB, pmrD mutations, and plasmid-borne mcr genes (Luo et al., 2017). Mobile genetic elements such as plasmids, transposons, and insertion elements accelerate the spread of β-lactamase resistance genes along this transmission chain, posing a significant threat to human health. Furthermore, there is an urgent need for systematically developed and regularly updated antibiotic treatment regimens tailored to different resistant bacteria.
3. Bacterial mobile genetic elements
It is worth noting that although different groups of incompatible plasmids have differences in strain origin and gene carriage, they share many similar genetic elements, such as insertion sequences, transposons, and integrons. Insertion sequences (IS), transposons, and integrons are discrete DNA segments that can transfer themselves or even carry surrounding resistance genes to other locations in the genome through a cut-and-paste mechanism. Initially, the main distinction between insertion sequences and transposons was thought to be the presence or absence of passenger genes, but as more gene elements have been detected, the boundaries between them have become increasingly blurred (Siguier et al., 2014). IS elements are primarily classified into 17 families based on their structural characteristics, including IS1, IS3, IS4, IS5, IS6, IS21, IS30, IS66, IS91, IS110, IS200/605, IS256, IS630, IS982, IS1380, ISAS1, and ISL3. Jacques Mahillon and Michael Chandler provided a comprehensive review of these families (Mahillon and Chandler, 1998). Two transposon families mainly associated with antibiotic resistance are Tn3 and Tn7-like transposons (Partridge et al., 2018). ISfinder (www-is.biotoul.fr) is a dedicated database for querying and annotating IS elements (Siguier et al., 2006). Furthermore, transposases can be classified into four types based on their structural features: DD(E/D) transposases, HUH single-strand DNA transposases, serine transposases, and tyrosine transposases. Integrons consist of three parts: the gene intl encoding the integrase, the gene attI encoding the recombination site, and the gene Pc encoding the promoter. Integrons are classified into types 1, 2, and 3 based on the similarity of their integrase genes, with type 1 integrons being frequently associated with microbial resistance (Bhat et al., 2023). These genetic elements not only help maintain the presence of resistance genes but also play an essential role in the horizontal transmission of plasmids. They facilitate plasmids in capturing exogenous genes and adapting to environmental changes, thereby promoting plasmid diversity and dissemination among different bacterial species. The presence of conjugative transfer regions in plasmids further enhances the spread of resistance genes. In these regions, there are regulatory factors and transfer genes that form a complex regulatory network.
4. The genetic structure of incompatible plasmids
Describing the biological characteristics of plasmids, which are crucial structures for bacterial resistance, is particularly important. Currently, plasmids of interest in Gram-negative bacteria are classified into 27 incompatibility groups, including IncA/C, B, F, H, I, L/M, K, P, T, O, X, W, R, Y, and their variants (Chen et al., 2024). These plasmids vary in size from several thousand to tens of thousands of base pairs (see Table 1 for the sizes of plasmids described in this article). They are extrachromosomal genetic material independent of the chromosome, consisting primarily of core and accessory genomes. The core genome refers to shared genomic sequences among all plasmids of this type, typically encompassing replication region, the maintenance region, and the conjugation transfer region. The replication region of plasmids contains rep genes, which control plasmid DNA synthesis and facilitate vertical transmission. Research also suggests that mutations in the antisense stem-loop RNA structure (RNAI) of incompatible plasmids affect the translation of rep mRNA, thereby altering plasmid compatibility (Rozwandowicz et al., 2023). Additionally, the conjugative transfer region contains tra genes, enabling horizontal transfer via the concerted action of the type IV secretion system (T4SS) and relaxase. The accessory genome comprises bacterial genome variable regions, primarily containing insertion sequences, transposases, integrons, antibiotic resistance determinant clusters, and heavy metal detoxification proteins. Through these elements, plasmids can capture new genes, and even mutations, endowing host bacteria with additional traits, thus enabling bacterial survival in more complex environments. Figure 1 displays the structural pattern of the plasmid. Mobile elements play a crucial driving role in the process of gene horizontal transfer. Insertion sequences, as part of transposons, act together with surrounding resistance genes to undergo horizontal transfer through a cut-and-paste mechanism, diversifying the genetic environment of resistance genes. This adaptation allows their hosts to cope with environmental challenges, potentially leading to the epidemic spread of a particular genotype of bacteria and the subsequent cross-species transmission (Macesic et al., 2023). Therefore, through in-depth research on the host distribution and geographical distribution of incompatible plasmids, conjugative transfer, carried resistance genes, and the function of genetic elements, we can have a more comprehensive understanding of the evolution process of bacterial resistance and provide more scientific guidance for tackling the problem of bacterial resistance.
Table 1.
Characteristics of plasmids and the commonly carried antibiotic resistance genes.
| Inc groups | Subtype | Size (bp) | β-lactamase resistance gene |
|---|---|---|---|
| IncA | A | 52637- 272299 | bla VIM-1, bla SHV-12 |
| IncC | C | 46056- 436531 | blaTEM-1B , bla CMY-2, bla NDM-1, bla CMY-4, bla CMY-6 , bla CMY-7, bla OXA-10 |
| IncF | FII: FIA: FIB | 2592- 583215 | bla CTX-M-15, bla CTX-M-14, bla CTX-M-27, bla KPC-2, bla SHV-12 |
| IncH | HI, HII | 50273- 477340 | bla CTX-M-9, bla TEM-1 , bla SHV-12 |
| IncL | L | 26406- 167203 | bla OXA-48, bla CTX-M-9 , bla CTX-M-17 |
| IncM | M1, M2 | 29895- 156910 | bla CTX-M-4, bla OXA-48 , bla CTX-M-9 , bla IMP-4 |
| IncP | P-α、-β、-γ、-δ、-ϵ、-ζ | 8187- 496601 | bla KPC-2 , bla OXA-2 |
Figure 1.
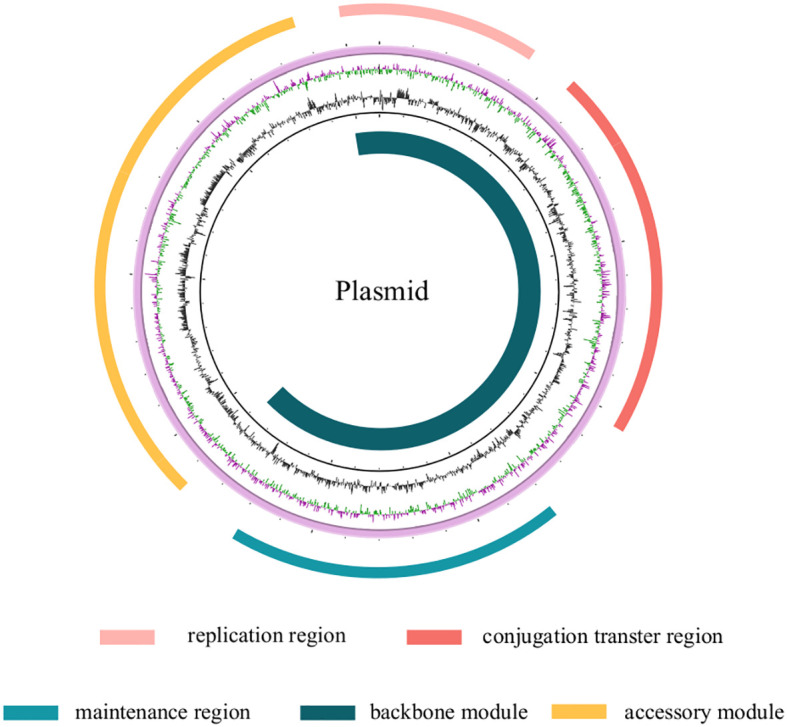
The plasmid genome is composed of two parts: the backbone module and the accessory module. The backbone module includes a replication region, a stability maintenance region, and a conjugation transfer region, while the accessory module is mainly comprised of antibiotic resistant genes. The two modules are not distinct from each other, but rather, they are interspersed.
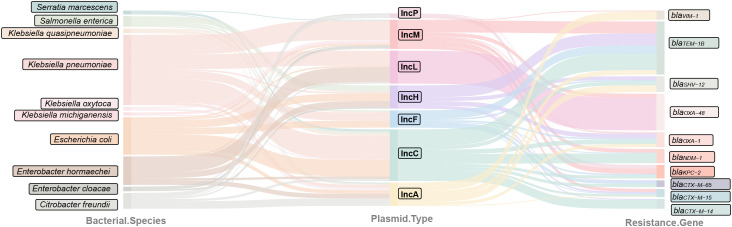
5. Distribution and association of incompatible plasmids with antibiotic resistance genes
5.1. IncA and IncC plasmid
The earliest occurrence of IncA and IncC plasmids may be associated with the use of antibiotics in animal husbandry (Johnson and Lang, 2012). The initial IncA plasmid was isolated from the fish pathogen Aeromonas hydrophila by Aoki et al. in 1971 and named pRA1 (accession number: FJ705807), later becoming the reference plasmid for IncA. The IncC plasmid was first discovered in the 1960s in Salmonella typhimurium, Pseudomonas aeruginosa (pIP40a), and Klebsiella pneumoniae, isolated from hospitals in Paris (Harmer and Hall, 2015). The historically significant IncC plasmid among them is the pR55, which was isolated from a strain of Klebsiella pneumoniae in 1969 and is also the oldest known IncC plasmid. The difference between IncA and IncC plasmids is the presence of a 69-nucleotide difference in the repA amplification sub-sequence, but they still share at least 98% identity. The genes necessary to perform replication (repA), conjugative transfer (tra), and plasmid partitioning (stb and par) make up the pRA1 plasmid’s backbone. The main difference between the pR55 and pRA1 plasmids lies in their accessory genes. The pR55 plasmid carries Tn6178, which contains the catA1, addB, bla OXA-21, and sul1 resistance genes, as well as the ISCR2 genetic element, which carries the sul2 and floR resistance genes. On the other hand, the pRA1 plasmid carries the sul2 and tet resistance genes (Doublet et al., 2012). Most IncC plasmids share a high degree of homology in their core genes, which include the essential genes for plasmid replication, distribution, stable maintenance, and conjugative transfer. In the early 1990s, Datta and Hedges, along with others, discovered that pRA1 (the reference plasmid for IncA) is compatible with plasmids from all known compatibility groups, including IncC plasmids. However, in 1972, Hedges observed that the pRA1 plasmid had significant entry exclusion from the IncC plasmid, so the IncA and IncC plasmids were called the “A-C complex”. IncA/C was later introduced and became popular. Ambrose et al. advised doing away with the labels IncA/C, IncA/C1, and IncA/C2 after demonstrating the compatibility of IncA and IncC replicons in 2018. They recommended referring to IncA/C as IncA and IncC individually or using “A/C,” “repA/C,” or “A-C complex” instead (Harmer et al., 2017; Ambrose et al., 2018). Specifically, IncA corresponds to IncA/C1, IncC to IncA/C2, and IncC comprises two types, C1 and C2. The primary distinction between C1 and C2 plasmids is that C1 is composed of orf1832 and rhs1, whereas C2 includes orf1847 and rhs2 along with two insertion sequences i1 and i2 (Harmer and Hall, 2015). Mi-Jung Kim and colleagues conducted sequencing and analysis of A/C plasmids extracted from photobacterium damselae subsp. piscicida strains in the United States (pP91278) and Japan (pP99-018) during the 1990s. They found that compared to the pP99-018 plasmid, the pP91278 plasmid had an additional 1kb resistance determinant cluster and a conjugative transfer region IV (TRA IV). The remaining regions of the two plasmids showed >90% identity, providing favorable evidence for horizontal transfer of plasmids between bacteria and further evolution (Kim et al., 2008).
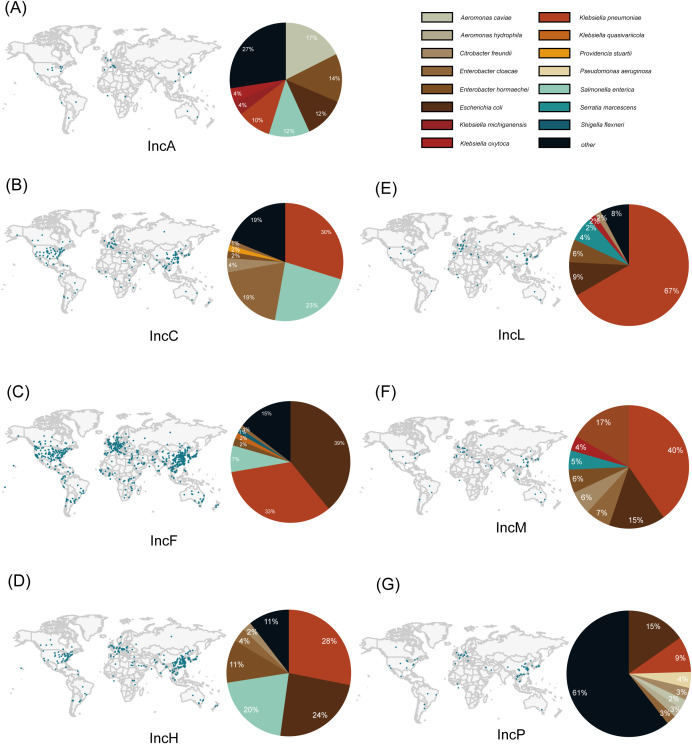
The IncA plasmid has been reported relatively infrequently, with a total of 51 cases distributed across the USA, Italy, China, Australia, the United Kingdom, Spain, Brazil, and other countries ( Figure 2A ). In contrast, the IncC plasmid has been reported more frequently, with a total of 831 cases distributed across 49 countries including the USA, China, India, Japan, and Germany. Among these countries, the United States and China have reported the highest number of IncC plasmids ( Table 2 ). It has been found that IncA plasmids are mainly present in the host bacteria Citrobacter freundii (n=9 cases) ( Figure 2A ), while IncC plasmids are primarily distributed in bacteria such as Klebsiella pneumoniae (n = 247), Salmonella enterica (n=192 cases), and Escherichia coli (n=161 cases) ( Supplementary Material 1 ) ( Figure 2B ). The specimens for isolation of IncA plasmids mainly comprise human perianal swabs, while IncC plasmids are predominantly sourced from human urine, blood, and perianal swabs. Additionally, isolation sources for both plasmids include strains from animals and the environment.
Figure 2.
Global distribution of plasmids and host bacterial distribution. The global distribution map is obtained from the PLSDB database (Galata et al., 2019; Schmartz et al., 2022). The pie chart represents the distribution of plasmid host bacteria, displaying the top six most abundant plasmid types, with the remaining grouped as “other”. (A–G) represent the geographic distribution and host bacterial distribution of IncA, IncC, IncF, IncH, IncL, IncM, and IncP plasmids, respectively.
Table 2.
Host bacterial information of plasmids and genetic environment of antibiotic resistance gene.
| Inc groups | Total | Major host bacteria | Major country | Antibiotic-resistant genetic environment | Accession number |
|---|---|---|---|---|---|
| IncA | 51 | Citrobacter freundii, Enterobacter hormaechei Escherichia coli, | USA, Italy, China | – | – |
| IncC | 831 | Klebsiella pneumoniae, Salmonella enterica Escherichia coli, | USA, China, Japan | ISEcp1-bla CMY-2 -bla-sugE | HQ023863 |
| HQ023862 | |||||
| FJ621588 | |||||
| ISEcp1-bla CMY-4 -blc-sugE | AJ704863 | ||||
| sugE-blc-bla CMY-2 -ISEcp1 | KU160531 | ||||
| sugE-bla CMY-2-1 -bla CMY-2-1 -sugE | CP000604 | ||||
| ISEcp1-bla CMY-6 | JN157804 | ||||
| IncF | 10,778 | Escherichia coli, Klebsiella pneumoniae, Salmonella enterica | China, USA, United Kingdom | fosA3-IS26-IS26-bla TEM-1-bla CTX-M-3-ISEcp1 | JX627737 |
| Tn3-like-ISEcp1-blaCTX-M-15-IS26-IS26 | GU371926 | ||||
| GU371929 | |||||
| fosA3-IS26-intI1-IS26-bla TEM-1-bla CTX-M-3-ISEcp1 | JQ432559 | ||||
| IS1-Tn2-IS26-intI1-dfr-qepA-ISCR3-groEL/intI1-rmtB-bla TEM-1-Tn2-IS26-Tn2-IS26-Tn21-IS26 | JX997935 | ||||
| IS26-intl1-dfr-qepA-ISCR3-groEL/intI1-rmtB-bla TEM-1-ltnpR-IS26 | AB263754 | ||||
| AM886293 | |||||
| FJ183463 | |||||
| FJ167861 | |||||
| IncH | 1,618 | Klebsiella pneumoniae, Escherichia coli, Salmonella enterica | China, USA, Japan | IS26-bla SHV-12 -IS26, IS26-catA2-IS26, IS26-tetR(D)-tetA(D)-IS26, IS26-catA2-IS26 | KY270852 |
| IS26-tetR(D)-tetA(D)-IS26, IS26-catA2-IS26 | KY270851 | ||||
| intI1-bla IMP-8 -aacA4-catB3-qacEdelta1-sul1-ISCR1-dfrA19-intl1-strA-strB | EU855787 | ||||
| intI1-bla IMP-8 -aacA4-catB3-qacEdelta1-sul1-ISCR1-qnrB2-sul1-ISCR1-dfrA19-intl1-strA-strB | EU855788 | ||||
| IncL | 407 | Klebsiella pneumoniae, Escherichia coli, Enterobacter hormaechei | Spain, Netherlands, China | Tn2-bla TEM-1 -tnpA-res-frmB | KM406489 |
| IS1999-lysR-bla OXA-48 -IS1-IS1999 | KM406491 | ||||
| IS1999-IS1R-bla OXA-48 -lysR-IS1999 | KC335143 | ||||
| IS1999-lysR-bla OXA-48 -IS1999 | JN626286 | ||||
| IncM | 179 | Klebsiella pneumoniae, Escherichia coli, Enterobacter cloacae | USA, China Australia | bla TEM-1 -tnpAcp2-aacC2-IS26-bla NDM-1 -trpF-bla DHA-1 -ampR-hypA-qac-sul1-ISCR1 | NC_019063 |
| bla TEM-1 - ISCfr1-aacC2-IS26-bla NDM-1 -ampR-sul1-ISCR1-ISEc28-armA-ISEc29-msrE-mphE-IS26-Tn2 | NC_019889 | ||||
| int1-blaIMP-1-aac (6’)-IIc-qacL-qacE-delta1-sul1-istB-IS21 | AP024913 | ||||
| IncP | 316 | Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa | China, Spain, Japan | aacA4-bla IMP-9-aacA4, aacA4-catB8a-bla OXA-10 | KC543497 |
| Intl1-aacA4-bla IMP-45-bla OXA-1-catB3-qacE1-sul1 (In786) | CP061377 | ||||
| Intl1-aacA4-bla IMP-45-guc35-bla OXA-1-catB3-qacED1-sul1(In786) | KY883660 |
A/C-type plasmids, found in various geographical locations and host organisms, share a similar plasmid backbone: genes related to replication, maintenance, regulation, DNA metabolism, and conjugation transfer. Furthermore, the backbone includes many potential DNA-binding transcriptional regulation factors (referred to as nuclear-associated proteins, NAPs) that can reduce the fitness cost and expand the host range of the plasmid population. Accessory modules of A/C-type plasmids are mainly composed of insertion sequences, Transposases, class 1 integrons, antibiotic resistance genes, and heavy metal detoxification proteins genes (Rada et al., 2022). IncA and IncC plasmids exhibit high degree of gene homology and share highly conserved gene backbones. The A/C plasmid backbones region can be divided into the replication region (repA), stability region (parAB, stbA, and a toxin-antitoxin system), and conjugative transfer region (aca, tra). The prominent features of the backbones are the replication origin and Type IV secretion-like conjugative transfer system of A/C plasmids. The smallest sequenced IncC plasmid to date, pKPHS2, still carries the conjugative transfer gene tra and 13 antibiotic resistance determinants. The IncC plasmid backbone also contains the dcm1, dcm2, and dcm3 genes, which encode DNA cytosine-5-methyltransferases and prevent degradation by host nucleases (Harmer and Hall, 2015; Ambrose et al., 2018). The reference plasmids for IncC type 1, pR148, and IncC type 2, pRMH760, have over 120 open reading frames in their backbone regions, encoding more than 100 amino acids (AAs). Toxin/antitoxin systems, DNA metabolism, conjugative transfer, partitioning, and stability, as well as several genes with unidentified roles, are among the genes necessary for replication (Del Castillo et al., 2013). The backbone regions of IncC plasmids of type 1 and type 2 differ by only 1%. This indicates that both types of plasmids have originated from a common ancestor and have undergone separate evolution in different lineages.
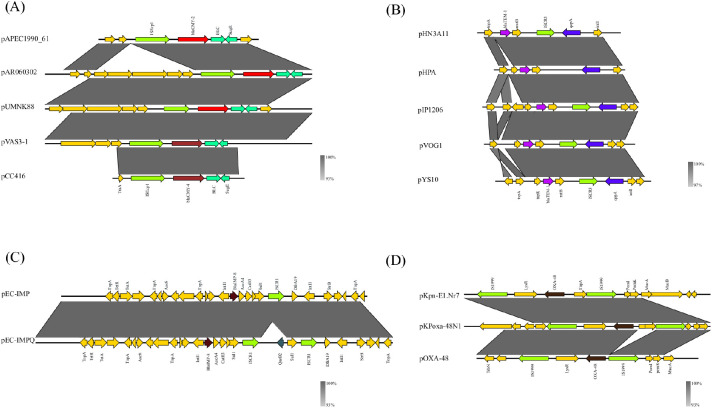
A/C plasmids are having a wide global distribution and commonly associated with multidrug resistances. Among them, IncA plasmids carry three potential resistance genes or clusters: sul3 (sulfonamide resistance gene), tetRA (class D tetracycline resistance gene cluster), and hipAB-related gene clusters. Plasmid pRA1 exhibits resistance specifically to sulfonamides and tetracyclines (Fricke et al., 2009). The IncA plasmid primarily harbors β-lactam resistance genes, including bla VIM, bla TEM-1B, and bla SHV-12, and is known for its broad host range. Among IncC plasmids, the carriage rate of the carbapenem-coding resistance gene bla NDM is the highest, with the bla NDM-1 subtype being particularly prevalent, which has emerged in IncC plasmids only recently. ( Figure 3 ). Additionally, it is common to find the bla CMY gene in IncC plasmids, with bla CMY-2 being the most predominant subtype, followed by bla CMY-7, and a small number of bla CMY-4 and bla CMY-6 subtypes. Additionally, the plasmids also carry carbapenem-coding resistance genes, primarily bla NDM-1, followed by bla KPC-2 and bla IMP-4. In research conducted by Christopher J. Harmer and colleagues, IncC plasmids containing both bla NDM-1 and bla CMY-2 are classified as Type 1, and all Type 1 IncC plasmids have ARI-A in the same location within their backbones. An array of antibiotic resistance genes can also be found in ARI-A, and both Type 1 and Type 2 IncC plasmids contain ARI-B. In IncC plasmids, the carriage rate of the carbapenem resistance gene bla NDM is the highest, particularly with the prevalence of the bla NDM-1 subtype. This gene has only started to appear in IncC plasmids in recent years. Statistics show that almost all IncC plasmids carry the sul1 or sul2 gene, indicating inherent resistance to sulfonamide drugs. Plasmids containing the bla CMY-2 gene are usually surrounded by insertion sequences or transposons, of which plasmids pAR060302, pUMNK88_161, and p199061_160 share the same gene environment with ISEcp1-bla CMY-2-blc-sugE ( Figure 4A ) ( Supplementary Material 3 ). Additionally, the plasmids pVAS3-1 carrying the bla CMY-4 gene and pCC416 share a genetic context identical to the aforementioned: ISEcp1-bla CMY-4-blc-sugE ( Table 2 ). Furthermore, the plasmid pNDM-KN carrying the bla CMY-6 gene also contains ISEcp1 genetic elements, with only one amino acid substitution (W661→L) compared to the bla CMY-2 gene (Carattoli et al., 2012; Partridge et al., 2018). The bla CMY-2 gene may potentially spread horizontally through the formation of a transposon unit (ISEcp1-bla CMY-2-blc-sugE), followed by recombination or mutation. In the process of interspecies transfer of resistance genes, the ISEcp1 genetic element plays a crucial role. Relevant studies indicate that ISEcp1 can provide a promoter for the captured gene at least, and it can mobilize adjacent DNA segments, which may explain the presence of bla CMY-2 and its variants in IncC plasmids. IncC carries resistance genes including bla TEM, bla SHV, bla CTX-M, bla DHA, bla OXA, bla IMP, sul1, sul2, aphA1, aadA, aadB, strA, strB, aacC, tetA, floR, catA1, and dfrA. Furthermore, on IncA/C plasmids isolated from Escherichia coli, the genes fosA3 and bla SFO-1 have been found (Zhao et al., 2015). There are differences in the resistance genes carried by Type 1 and Type 2 IncC plasmids. The bla TEM-1 gene is more likely to be present in Type 2 IncC plasmids, while the bla CMY-59 gene is mainly distributed in Type 1 (Zhang et al., 2022). In addition, certain A/C plasmids also contain resistance genes associated with quaternary ammonium compounds (qacE and sugE) and mercury-related resistance genes, such as pSH163_135 (accession number: JN983045) and pSH696_135 (accession number: JN983048) (Han et al., 2012).
Figure 3.
Relationships between plasmids, host bacteria, and β-lactam resistance genes. The data were sourced from the latest 50 discoveries of each plasmid type in the PLSDB database, with the top 10 host bacteria and genes by quantity used for the diagram.
Figure 4.
Genetic environment of plasmids carrying antibiotic resistance genes. (A) IncC plasmids: the genetic environment of blaCMY-2 in pAPEC1990_61, pAR060302, and pUMNK88, and of blaCMY-4 in pVAS3-1 and pCC416. (B) IncF plasmids: the genetic environment of blaTEM-1 in pHN3A11, pHPA, pIP1206, pVOG1, and pYS10. (C) IncH plasmids: the genetic environment of blaIMP in pEC-IMP and pEC-IMPQ plasmids. (D) IncL/M plasmids: the genetic environment of blaOXA-48 in pKpn-E1, Nr7, pKPoxa-48N1, and pOXA-48.
Compared to mutations occurring within plasmids themselves, inter-strain plasmid transfer contributes more to the survival of bacterial populations. However, the transfer frequency is mainly influenced by factors such as recipient bacteria, donor bacteria, compatibility of resident plasmids, and experimental conditions for conjugation. Although transfer frequencies may be high, they do not necessarily confer a survival advantage to bacterial populations (Geoffroy and Uecker, 2023). A/C plasmids belong to transferable plasmids, but different A/C plasmids exhibit varying transfer frequencies. Under natural conditions, the transfer frequency of IncA plasmid pRA1 in solid culture is 10-2, while in broth it is 10-4, and the transfer frequency of IncC plasmids ranges from 10-7 to 10-4 in both solid and broth cultures (Harmer and Hall, 2015). In comparison, the transfer frequency of IncC plasmids is higher than that of IncF plasmids. Factors controlling the transfer ability of IncC plasmids appear to be related to the chromosome. Genes involved in the conjugative transfer mechanism of A/C plasmids are mainly tra genes and mobI genes (Carraro et al., 2014). The conjugation transfer genes in the A/C plasmid have been identified by matching the protein to the known Tra protein; the IncA and IncC groups also have analogous genes. The transfer genes are divided into two distinct regions. The first region contains three sets of genes, which were isolated through seven unknown functional reads. The first group consists of traI and traD, the second group consists of traL, traE, traK, traB, traV, and traA, and the third group consists of traC, trhF, traW, traU, and traN. The final three transfer genes—traF, traH, and traG—are located in the second region. These genes produce mating pair stabilizing proteins, relaxation enzymes from the MOBH group, coupling proteins, and proteins from the Mating pair formation (MPF) group that are essential for the assembly of the Type IV secretory system (T4SS), among other proteins involved in the conjugative processes. AcaCD, a transcription activator complex, is essential for the conjugative transfer of IncC plasmids. Its binding location is upstream of traI, traL, traV, traN, and traF, and it promotes the transcription of the tra genes. In addition, AcaCD also boosts the expression of 14 more IncC backbone areas, and potential AcaCD binding sites have been found in each of these regions (Gordils-Valentin et al., 2024). Therefore, its conjugative transfer frequency is influenced by multiple genes.
The IncA plasmid and the IncC plasmid are relatively small, and both can undergo conjugative transfer. However, the IncC plasmid is more abundant than the IncA plasmid, which may be related to the broader host range of IncC. The most common host for the IncC plasmid is Klebsiella pneumoniae, which has a more widespread distribution in nature. Currently, carbapenems are commonly used to treat infections caused by Gram-negative bacteria. The gene encoding carbapenemase, bla VIM, is frequently carried by the IncA plasmid, which raises the need for heightened vigilance.
5.2. IncF plasmid
The IncF plasmid was the first antibiotic resistance plasmid discovered in bacteria and is currently the most reported plasmid type. This plasmid can carry multiple replicons simultaneously, defining the FII, FIA, and FIB replicons based on the differences in copA, repA, and repB gene sequences. In 1960, Rintaro Nakaya and his colleagues isolated an IncF plasmid from Shigella flexneri in Japan, which belonged to the FII replicon group. Two years later, Yoshinobu Sugino and Yukinori Hirota proposed the association of this plasmid with antibiotic resistance (Sugino and Hirota, 1962). Most of the IncFI plasmids were isolated from the 1970s. In 2008, Bruno Pe´richon and others identified the IncF plasmid pIP1206 in Escherichia coli in France and conducted whole-genome sequencing. This plasmid has four replication regions: repFIA, repFIB, repFIIA, and repFIIB. The repFIA replication region of the pIP1206 plasmid is 99% identical to that of the pRSB107 plasmid. At the same time, the repFIB replication region shows a 99% consistency with the replication regions of both pRSB107 and pAPEC-O1-ColBM plasmids. The authors also proposed the viewpoint that pIP1206 is likely a new plasmid formed by recombination of pRSB107 and a pAPEC-O1-ColBM-like plasmid (Périchon et al., 2008). This indicates that plasmids can undergo recombination, similar to genes, and form new plasmids when their replicons are identical. This phenomenon allows for the bundling of resistance genes carried by plasmids, resulting in multiple resistance genes appearing on the same plasmid, thus conferring a higher level of drug resistance to the host bacteria. In 2010, it was first reported that an IncF plasmid carrying bla TEM-1, bla SHV-12, and bla CTX-M-15 genes was isolated from Klebsiella pneumoniae strains, which facilitated the simultaneous dissemination of these resistance genes (García-Fernández et al., 2010).
A total of 10,778 cases of IncF plasmids have been reported, with the highest prevalence found in Escherichia coli (n=4,287 cases), followed by Klebsiella pneumoniae (n=3,666 cases), Salmonella enterica (n=777 cases), Enterobacter hormaechei (n=221 cases), and Klebsiella quasivariicola (n=166 cases) ( Table 2 ). IncF plasmids have been found in almost every country, with a prevalent distribution observed in countries such as China, USA, the United Kingdom, Spain, Japan and Australia ( Supplementary Material 1 ) ( Figure 2C ). Samples from a diverse range of hosts have been identified as carriers of IncF plasmids, including humans, animals, and water sources. Within human samples, blood is the most common and significant source, followed by feces, urine, and sputum. Some IncF plasmids have also been found in abdominal drainage fluid, pus, bile, and perianal swabs.
The IncF plasmid is characterized by its large size and exhibits a complex mosaic structure. Among the multiple replicons mentioned above, the FII replicon is the most common type, followed by the FIB replicon. Additionally, IncF plasmids typically have the ability to carry multiple replicons, thereby expanding their host range (Ammar et al., 2024). Based on the typical replicons FII, FIA, and FIB, a replicon sequence typing (RST) scheme has been proposed, represented using the FAB formula. For example, F1:A1: B-, where F represents the IncFII replicon, A represents the IncFIA replicon, and B represents the IncFIB replicon. According to our data analysis, F2: A-: B- plasmids are the most prevalent, and the majority of them carry the antibiotic resistance gene bla CTX-M-15. Clearly, this plasmid formula has become the major carrier for spreading antibiotic resistance genes. The backbone region of IncF plasmids mainly includes replication-related genes (repFIA, repFIB, repFII, etc.), plasmid maintenance genes (toxin-antitoxin systems such as type I: hok-sok, type II: relBE, mazEF, vapBC, ccdAB, parDE, higAB, HipBA, and Phd-Doc), and conjugative transfer genes (tra, trb, artA, and finO). Additionally, certain IncF plasmids also carry exclusion genes (traS and traT) responsible for plasmid incompatibility (Oluwadare et al., 2020). The accessory module contains plasmid resistance genes, cargo genes (such as ompP, ychA, and ychB), and virulence genes (Al Mamun et al., 2021).
IncF plasmids are most commonly associated with β-lactamase resistance genes, the highest carriage rate of which is observed for bla TEM-1B, followed by bla CTX-M, bla SHV, and bla KPC( Figure 3 ). The subtypes of bla CTX-M found on IncF plasmids mainly include bla CTX-M-1, bla CTX-M-3, bla CTX-M-9, bla CTX-M-14, bla CTX-M-15, bla CTX-M-27, and bla CTX-M-55. Among these, the most common subtype is bla CTX-M-15, which is consistent with the findings of Yasufumi Matsumura et al (Matsumura et al., 2013). The most commonly found carbapenemase-resistant gene is bla KPC-2. Interestingly, we observed that, although the resistance genes carried by IncF plasmids and IncC plasmids do not completely overlap, there are ISEcp1 gene elements upstream or downstream of the carried resistance genes. For example, in plasmids pEC_B24, pEC_L8, and pEC_L46, the resistance gene bla CTX-M-15 is present, and the surrounding gene environment contains the ISEcp1 element. Additionally, the sequence ISEcp1-bla CTX-M-15 on plasmids pEC_B24 and pEC_L46 is located on a Tn3-like transposon, specifically within the Tn3-like–ISEcp1-bla CTX-M-15-IS26-IS26 sequence ( Supplementary Material 3 ). IS26 plays a crucial role in gene recombination (Vázquez et al., 2021). A similar structure exists in plasmids identified by Xiaojie Chen et al., including pHN3A11 (accession number: JX997935), pHPA (accession number: AB263754), pYS10 (accession number: FJ167861), pVOG1 (accession number: FJ183463), and pIP1206 (accession number: AM886293), where a conserved sequence of IS26-Δintl1-dfr-qepA-ISCR3-groE/Δintl1-rmtB-bla TEM-1-ItnpR-IS26 is present ( Figure 4B ). This indicates the significant role of IS26 and ISCR3 should not be underestimated (Chen et al., 2014). The Tn3-like transposon, as a mature transposable element, facilitates the transfer of the bla CTX-M-15 resistance gene between plasmids or hosts, which could potentially explain its widespread presence in IncF plasmids. The fact that the ISEcp1 element appears not just upstream or downstream of bla CTX-M-15 but also upstream of bla CTX-M-14 suggests that it plays a significant function in the neighborhood of resistance genes in general (Dhanji et al., 2011). Research indicates that within the same bacterial species, there is a high degree of identity in the mobile element regions carried by plasmids. According to Ling Wang’s study, the 18.9 kb IS26 composite transposon carried by IncF plasmids in Escherichia coli shows an identity rate as high as 97.4%. Additionally, apart from IS26 and ISEcp1, IncF plasmids are frequently associated with mobile genetic elements such as IS1, IS6100, IS6, IS903, ISKpn6, ISKpn27, Tn3-like, Tn1712, In27, and In54 (Gancz et al., 2021; Hounmanou et al., 2021). Other types of resistance genes carried by IncF plasmids include: bla NDM-1, bla NDM-5, bla NDM-7, bla NDM-11, bla KPC-3, bla KPC-8, bla OXA-1, bla OXA-3, bla OXA-48, bla OXA-181, bla CMY-2, bla TEM-1, bla GES-5, mcr-1, mcr-9, armA, arr-3, rmtB, ΔqacE, qeqA, qnrS, qnrB, aadA2, aadA5, dfrA7, catB4, aac(6’)-Ib-cr, aph(3’)-Ib, aph(6’)-1d, acc (3’)-IId, floR, mph(A), sul1, sul2, strA-strB, and heavy metal resistance genes such as copper and silver (Stephens et al., 2020; Conte et al., 2022; Liang et al., 2022; Turton et al., 2022). Furthermore, it has been observed that the resistance gene aac (6’)-Ib-cr frequently co-occurs with bla CTX-M-15 or bla TEM-1, and bla NDM-5 is commonly found together with rmtB on IncF plasmids, which will further exacerbate the situation of multidrug resistance. Over time, an increasing number of new subtypes of resistance genes have emerged in plasmids. Juliana Buck Dias et al. were the first to identify an IncF plasmid carrying the bla CTX-M-24 gene in Escherichia coli, and ShiKai Wu et al. also identified an IncF plasmid carrying the bla OXA-1041 gene. This resistance gene shows high similarity to bla OXA-917 and bla OXA-427 ( Dias et al., 2022; Wu et al., 2023). It is evident that the resistance spectrum of IncF plasmids is becoming increasingly broad, which implies the existence of mutations, transfers, or acquisitions of resistance genes. It is worth delving deeper into the mechanisms behind this phenomenon in order to alleviate the problem of the lack of effective drugs in clinical settings due to resistance.
The IncF plasmid’s conjugation transfer function is mainly regulated by three genes: traM, traJ, and traY. traM is involved in DNA processing and controls its own promoter, Pm . traJ is the activator for the tra gene, while traY-X helps regulate DNA metabolism, T4SS, and metastasis. traM activates Py to create traY, which then stimulates Pm and further boosts the expression of traJ. This causes the expression of the transfer Operon in the IncF plasmid, which leads to the production of relaxants (traD, traI, traM, traY), conjugated pili (traA, traQ, traX, traP, traE, traL, traC, traW, trbC, traF, trbB), pairing stable proteins (traN, traG, traU), and T4SS core proteins (traB, traK, traV), thus facilitating conjugation (Bragagnolo et al., 2020). As the concentrations of TraM and TraY go up, they will impede Pm and Py . TraM, TraJ, and TraY form a regulatory loop that is stimulated by the protein SfrA (ArcA) and IHF, which affect the promoter Py in the IncF plasmid. The negative regulator, Antisense RNA (finP), can suppress the translation of traJ mRNA. Moreover, conjugation is linked to the entry of TraS and TraT, whose expression is managed by traJ. When TraS binds to TraG, it hinders the conjugation transfer (Frost and Koraimann, 2010).
The IncF plasmid, as the most abundant plasmid type, is widely distributed among various bacterial strains, with Klebsiella pneumoniae being the most common. It carries a rich reservoir of resistance genes and is prevalent in many countries. Additionally, its more complex plasmid structure poses a significant threat to human health.
5.3. IncH plasmid
IncH plasmids fall into one of two categories: IncHI and IncHII, with IncHI being the main type. Based on the repertoire of replication genes, IncHI plasmids can further be categorized into five different subtypes: IncHI1 through IncHI5 (Liang et al., 2017). In 1961, the IncHI plasmid (R27: accession number AF250878) was first identified in Salmonella typhi in the United Kingdom. This plasmid carries the Tn10 transposon, which contains the tetracycline resistance gene. Similarly, in 1993, an IncHI plasmid (pHCM1) was isolated from Salmonella typhi CT18. The pHCM1 plasmid showed more than 99% sequence identity with the R27 plasmid and carries additional resistance determinants, including dhfr1b, cat, mer, strAB, sulII, and bla TEM-1 ( Sherburne et al., 2000; Parkhill et al., 2001). In 1969, Antone A. Medeiros et al. discovered the prototype IncHI2 plasmid, R478 (accession number: BX664015), in Serratia marcescens in the United States. The complete nucleotide sequence of this plasmid was later sequenced by Matthew W. Gilmour et al. in 2004 (Gilmour et al., 2004). The R478 plasmid carries a cluster of resistance determinants including tetracycline (also located on the Tn10 transposon), chloramphenicol, kanamycin, mercury, silver, copper, arsenic, and tellurite resistance genes. It is evident that the R478 and pHCM1 plasmids carry enriched resistance genes compared to the R27 plasmid. The R478 and pHCM1 plasmids may have captured additional resistance genes on the basis of the R27 plasmid (with the presence of insertion elements IS26, IS1, and the transposon Tn10 providing more opportunities for plasmid-mediated acquisition of exogenous genes) and evolved from there. In 1985, E. Chaslus-Dancla et al. identified IncH plasmids in Escherichia coli. Subsequently, Timothy J. Johnson et al. first completed the sequencing of the IncHI2 plasmid (pAPEC-O1-R) isolated from E. coli in 2006, which shares a high similarity with the R478 plasmid (Chaslus-Dancla and Lafont, 1985; Johnson et al., 2006). IncHI2 plasmids were identified in a pneumonic Klebsiella pneumoniae (pK29) collected in 2001 and in two sewer Escherichia coli (pEC-IMP and pEC-IMPQ) collected in 2004 by Ying-Tsong Chen et al (Chen et al., 2007; Chen et al., 2009). Compared to plasmid R478, plasmid pK29 has two unique regions. One region contains the IS26-ISCR1-bla CMY-8-qacEdelta1-sul1-IS26-IS6100 sequence, and the other region contains the tnpA-bla CTX-M-3-tnpA sequence. The main difference between plasmids pEC-IMP and pEC-IMPQ is the presence of the qnrB2 resistance gene, which is also associated with different host resistance phenotypes ( Table 2 ). The genetic environment of the resistance genes in pEC-IMP and pEC-IMPQ plasmids are “intI1-bla IMP-8-aacA4-catB3-qacEdelta1-sul1-ISCR1-dfrA19-intl1-strA-strB” and “intI1-bla IMP-8-aacA4-catB3-qacEdelta1-sul1-ISCR1-qnrB2-sul1-ISCR1-dfrA19-intl1-strA-strB” respectively ( Figure 4C ). It is evident that the upstream ISCR2 gene element on qnrB2 plays a significant role in the horizontal transmission of the qnrB2 resistance gene.
A total of 1,618 cases of IncH plasmids have been identified, predominantly found in China, USA, the United Kingdom, Japan, Australia, Spain, South Korea, Canada, Switzerland and the Czech Republic. There are a total of 49 different host bacteria, with Klebsiella pneumoniae (n=454 cases), Escherichia coli (n=390 cases), and Salmonella enterica (n=329 cases) being the most commonly associated strains with this plasmid ( Supplementary Material 1 ) ( Figure 2D ). The main isolation sources of the specimens are human blood, feces, sputum, and urine, while in animals, they are primarily isolated from cattle and chickens.
The backbone region of the IncH plasmid contains replicative genes (repHI1A, repHI1B, and repHI2 for IncHI plasmid replication, with five different subtypes, each containing specific replicative genes: IncHI1: repHI1A+repHI1B+repFIA-like, IncHI2: repHI2A+repHI2C, IncHI3: repHI3B+repFIB-like, IncHI4: repHI4A+repHI4B, and IncHI5: repHI5B+repFIB-like) ( Liang et al., 2017). It also includes the partitioning modules (Par1 and Par2), maintenance genes, and conjugative transfer genes (the tra1 and tra2 regions). Additionally, accessory modules contain resistance determinants and several mobile elements.
The IncH plasmid carries several β-lactamase genes, such as bla TEM-1, bla SHV-12, bla CTX-M-1, bla CTX-M-8, bla CTX-M-9, bla SHV-12, bla IMP-8, bla NDM-1, and bla OXA-10( Supplementary Material 1 ). Among the β-lactamase resistance genes, bla CTX-M is widely observed, and its most common subtype is bla CTX-M-9. The bla CTX-M-15 subtype, which is known to have a complex genetic environment, was identified in an IncH plasmid named pENVA by Andreas Schlüter et al (Schlüter et al., 2014). The genetic environment surrounding bla CTX-M-15 consists of IS26-aacC2-bla TEM-1-tnpRATn2-ISEcp1-bla CTX-M-15-tnp-Tn2. Bla TEM-1 is located on the transposon Tn2, while bla CTX-M-15 is located on the transposon Tn2012. Interestingly, upstream of bla CTX-M-15, there exists a genetic element called ISEcp1, which is likely responsible for mobilizing the bla CTX-M-15 gene and integrating it into the Tn2 transposon that contains bla TEM-1. In recent years, there has been a continuous increase in IncH plasmids carrying the bla CTX-M-15 subtype. These IncH plasmids also carry other resistance genes, including tet, dfrA5, sul1, sul2, strA, strB, qnrB, mcr-1, mcr-3, mcr-9, aacC2, aadA, aphA1ab, catA1, as well as heavy metal resistance genes (mercury, copper, silver, arsenic, and tellurium), and others (Fukuda et al., 2022; Wang et al., 2024).
The transfer of IncHI is mainly directed and expressed by tra1 and tra2 regions. These regions contain genes that code for oriT transfer initiation, relaxation bodies (traI), conjugating proteins (TraG), and Mpf proteins (TrhY, TrhR, TrhF, TrhH, and TrhG) ( Lawley et al., 2002). The tra2 region (trhA, trhL, trhE, trhK, trhB, trhV, trhC, trhP, trhW, trhU, and trhN) is responsible for encoding the Mpf protein and synthesizing H pili. The trhO, trhZ, and htdA genes can control the rate of metastasis, with trhO and trhZ increasing the frequency of metastasis and htdA having the opposite effect. The Mating pair formation (Mpf) apparatus is composed of pili protein subunits and pili protein processing proteins, which are connected to recipient bacteria to transfer plasmid DNA (Lawley et al., 2003). Research also indicates that in salmonella typhi, the conjugative transfer of IncHI plasmids is influenced by temperature. Between 22-30°C, approximately 10-80% of recipient bacteria can acquire IncHI1 plasmids, whereas at 37°C, almost no recipient bacteria acquire the plasmid. This phenomenon is primarily related to two mechanisms: first, the expression of transfer genes is inhibited or the transfer genes are unstable, and second, at 37°C, the Mpf apparatus becomes inactive. The temperature sensitivity of IncHI1 plasmid transfer confers an advantage for the dissemination of antibiotic resistance genes in environmental pathogens (Taylor, 2009).
Similarly, the IncH plasmid is also a broad-host-range plasmid, and it is relatively large. Clinically, colistin has been reintroduced for the treatment of infections caused by Gram-negative bacteria and is often used as a last-resort antibiotic. Genes associated with colistin resistance, such as mcr-1 and mcr-9, are frequently linked to the IncH plasmid (Saavedra et al., 2017; Osei Sekyere et al., 2020). Moreover, this plasmid is associated with multiple β-lactam resistance genes and has been detected with the highest frequency in China, which may be related to the relatively lenient antibiotic usage policies in the country.
5.4. IncL plasmid and IncM plasmid
In 1970, Y. A. Chabbert and colleagues identified the IncM plasmid (R69) from Salmonella paratyphi and Klebsiella pneumoniae strains in France. This was the earliest discovery of such plasmids, and R69 carries the resistance genes bla TEM-1, tetA(B), aphA-1, and merA ( Chabbert et al., 1972). In 1973, Hedges and R. W. and their colleagues identified the IncL and IncM plasmids in Escherichia coli and classified them into separate groups. However, in 1978, Richards and Datta merged them and named them IncL/M (Hedges et al., 1974). Following the latest PBRT subtyping scheme proposed in 2015, Carattoli, A. et al. classified it into three different types: IncL, IncM1, and IncM2. In 1977, Richards, H. and Datta, N. identified the presence of an IncL plasmid (R471) in a strain of Shigella flexneri isolated from the United States. The plasmid was found to harbor the bla TEM-1 and merA resistance gene clusters (Carattoli et al., 2015). Although both R69 and R471 plasmids harbor the bla TEM-1 resistance gene, they have completely different genetic environments. Twenty-eight years later, Park, Yeon-Joon et al. also discovered IncL/M plasmids in Shigella flexneri strains in South Korea. However, the resistance genes carried by this type of plasmid are completely different from the resistance gene cluster in R471, including bla CTX-M-3, bla CTX-M-12, bla OXA-1, aac (6’)-lb-cr, and armA. During this time period, IncL/M plasmids may have undergone significant genetic evolution. Park, Yeon-Joon stated that the transfer rates of the plasmids carrying ISCR1, aac (6’)-Ib-cr, armA, and bla OXA-1 were 100%, and there is a close association between the horizontal dissemination of bla CTX-M resistance genes and the ISEcp1 genetic element (Park et al., 2009). Evidence once more affirms that ISCR1 and ISEcp1 are able to facilitate the horizontal dispersal of resistance genes by enabling the mobilization of resistance genes during conjugative transfer.
There are a total of 407 cases of IncL plasmids, with Klebsiella pneumoniae (n=273 cases) being the most common host bacterium ( Figure 2E ). These plasmids are primarily distributed in the Spain, Netherlands and China. IncM plasmids, on the other hand, have a total of 179 cases, with 94 cases of IncM1 and 85 cases of IncM2. The distribution of these two types of plasmids is relatively even. The most common host bacteria for them are Klebsiella pneumoniae (n=72 cases) and Escherichia coli (n=27 cases) ( Figure 2F ). Literature suggests that IncL/M plasmids mainly originate from Enterobacteriaceae in the Mediterranean region and Western Europe (Adamczuk et al., 2015). The primary isolation sources for the IncL plasmid in human specimens are urine, blood, and peritoneal drainage fluid. Additionally, besides these sources, the IncM plasmid is predominantly isolated from human perianal swab samples.
Although the backbones of IncL and IncM are different, they still share some common backbone region genes, including replication genes (repA), maintenance genes (parA, parB), the tra locus (tra), and some plasmid backbone stability genes including pemI, pemK, mucA, mucB, resD, and the transfer gene trb. Since plasmid pEL60 (accession number AY422214) does not carry any resistance determinant clusters or insertion sequences, it is considered to have the most typical IncL/M plasmid backbone genes. pEL60 only contains replication genes (rep), transfer genes (trb), stability genes (pemI, pemK, parA, parB), and the tra locus. In terms of maintenance genes, the importance of parAB genes is higher than that of pemIK genes (Mierzejewska et al., 2007). IncL and IncM plasmids both have exclusion genes (excA, traY) and relaxase genes (traX), although their sequences are not the same. Additionally, both IncL and IncM accessory modules include resistance genes and transposon elements.
There are still differences in the resistance genes present in the accessory modules of IncL and IncM plasmids. The β-lactam resistance gene predominantly carried by IncL plasmids is bla OXA-48, accounting for 88% of all IncL plasmids carrying resistance genes ( Figure 3 ). Moreover, plasmids carrying bla OXA-48 alone are the most common among IncL/M plasmids, such as pRAYY (accession number: KX524525.1), pEC745 (accession number: CP015075.2), pKpn-E1. Nr7 (accession number: KM406491.1), and pKPoxa-48N1 (accession number: KC757416.2), all of which contain bla OXA-48 located on the transposon Tn1999 ( Figure 4D ). The gene sequences upstream and downstream of Tn1999 show a similarity of over 90%. Additionally, IncL/M plasmids isolated from patient samples by Linda Hadjadj and Nadim Cassir also carry Tn1999. This indicates that bla OXA-48 can stably exist on IncL plasmids and can be rapidly spread (Mierzejewska et al., 2007). In addition, other β-lactam resistance genes that co-occur with bla OXA-48 on the same plasmid include bla CTX-M. Other β-lactam resistance genes carried by IncL plasmids include bla SHV, bla TEM, bla VIM, and bla KPC. Other identified resistance genes include sul1, merA, and so on. We found differences in the β-lactam resistance genes carried by IncM1 and IncM2. bla SHV and bla NDM are only present in IncM1 plasmids, mainly the bla SHV-30 subtype. The most common β-lactam resistance gene in IncM1 plasmids is bla OXA, followed by bla CTX-M, while in IncM2 plasmids, the most common gene is bla TEM, followed by bla IMP. Other resistance genes carried by IncM1 and IncM2 include tetA (AB), aacAC, aadA, merA, sul1, armA, dfrA12, mph (AE), qacG, qnrB2, catB3, as well as resistance genes for heavy metals (mercury, cobalt, zinc, and cadmium) (Adamczuk et al., 2015). Enterobacter cloacae, Klebsiella aerogenes, and Serratia marcescens were recently isolated from a single patient specimen and were found to harbor the bla IMP-1 resistance determinant cluster on an IncL/M plasmid designated as pSL264. The bla IMP-1 resistance gene carried by this plasmid exhibits a distinctive genetic environment structure, which is depicted as int1-bla IMP-1-aac (6’)-IIc-qacL-qacE-delta1-sul1-istB-IS21. Furthermore, conjugation transfer experiments involving this plasmid have demonstrated the horizontal transmission of the resistance genes it carries (Mori et al., 2021). Additionally, a clinical strain of Klebsiella pneumoniae yielded an IncL/M plasmid termed pCTX-M360, which carries the bla CTX-M-3 gene. pCTX-M360, in contrast to pEL60 devoid of resistance genes or transposon elements, exhibits variations solely attributed to the presence of the ISEcp1 gene element and Tn2 transposon harboring the bla CTX-M-3 gene. Therefore, it can be inferred that the emergence of the pCTX-M360 plasmid involved multiple insertion events rooted in the pEL60 plasmid (Zhu et al., 2009). Plasmids not only act as carriers for transferring between bacterial species, but they also consistently acquire exogenous gene elements, thereby imparting additional characteristics to the plasmid.
The conjugative transfer genes of the IncL/M plasmid include trbA, trbB, trbC, nikA, mobA, traH, traI, traJ, traM, traN, traO, traQ, traU, traW, and traY, but their specific functions are still unclear. The expression of the tir gene in the IncL plasmid inhibits conjugative transfer, reducing the frequency of bacterial conjugation (Potron et al., 2014).
The gene encoding carbapenemase, bla OXA-48, is primarily located on the IncL plasmid, which shares a similar genetic background. Tn1999 may play a crucial role in the dissemination of this resistance gene. In contrast, the IncM plasmid is mainly associated with the bla VIM gene. The host bacteria for both plasmid types are predominantly Klebsiella pneumoniae and Enterobacter hormaechei, both of which are broad-host-range plasmids capable of conjugative transfer.
5.5. IncP plasmid
The IncP plasmid is called IncP-1 in the genus Pseudomonas and IncP in the family Enterobacteriaceae ( Yakobson and Guiney, 1983; Adamczyk and Jagura-Burdzy, 2003). This plasmid can be classified into six subtypes: IncP-α, -β, -γ, -δ, -ϵ, and -ζ. However, as more IncP/IncP-1 plasmids are identified and typing methods become more refined, additional subtypes such as η, θ, ι, κ, o, λ, and μ have also emerged gradually (Popowska and Krawczyk-Balska, 2013; Hayakawa et al., 2022). In 1969, the research team of Lowbury et al. isolated a strain of carbencillin-resistant Pseudomonas aeruginosa from the blood samples of patients in the United Kingdom, revealing the existence of the IncP-1 plasmid for the first time (Lowbury et al., 1969). The classical IncP-β subtype plasmids, R751 and pB8, were discovered in Enterobacter aerogenes in 1974 and Pseudomonas sp. in 2000, respectively. These two plasmids have highly similar backbone modules, with differences primarily focused on the accessory modules. R751 contains Tn402, with a structure of intl1-dhfrllc-orfD-qacE. In comparison, pB8 carries a transposon derivative of Tn402, containing the bla OXA-2 resistance determinant cluster, with a structure of intl1-bla OXA-2-aadA4-qacE△1-sul1-orf5. The sequences on both sides are completely identical to those in R751, suggesting that these two plasmids might have originated from a common ancestor (Schlüter et al., 2005). In Vibrio alginolyticus, an IncC plasmid with the bla CMY-2 resistance gene was found, as well as an IncP plasmid with the bla VIM-1 resistance gene. The bla VIM-1 gene was likely acquired by the class I integron and inserted into the integration hotspot of the IncP plasmid, which is different from the IncC plasmid (Zheng et al., 2019). IncP plasmids have been found not only in human samples but also in the natural environment. In 2013, del Castillo et al. identified an IncP-1 plasmid (pP9014) in a strain of Photobacterium damselae subsp. piscicida that was sourced from fish for the first time. This plasmid carries genes tetRD, catII, and a β-lactam resistance gene. The gene environment structure sequence is as follows: bla-IS5-catII-tetR-D-IS26-Tn3 ( del Castillo et al., 2013). In 2021, Yusuke Ota et al. for the first time discovered IncP-1 plasmids harboring the bla KPC-2 resistance gene in strains of Citrobacter freundii (p5_CFTMDU) and Klebsiella variicola (p3_KVTMDU). The similarity between the p5_CFTMDU and p3_KVTMDU plasmids reached 99.8%. Both plasmids harbor the bla KPC-2 resistance gene in the genetic environment of ISKpn6-bla KPC-2-Tn3 (Ota et al., 2022). These plasmids, including pP9014, p5_CFTMDU, and p3_KVTMDU, highlight the ability of plasmids to serve as carriers for gene transfer between bacteria and natural environments. Moreover, the transposon Tn3 has become a common genetic element in the dissemination of antibiotic resistance genes.
A total of 316 IncP plasmids have been found in 24 countries, includingChina, Spain, Japan, USA, Viet Nam, Laos, Canada, Czech Republic, France, Germany, Argentina, Brazil, United Kingdom, Belgium, Denmark, Iran, Australia, Croatia, Ghana, Mexico, Missing, Netherlands, and South Korea. These plasmids have been detected in over 60 different host bacteria, with the most frequent one being Escherichia coli (n=49 cases) ( Figure 2G ). It is shown that the isolated source of IncP plasmid is relatively evenly distributed in human, environmental and animal samples, but unlike the other plasmids, it is more common as an isolated source in environments such as soil or sewage.
The backbone genes of IncP plasmids include the replication region, stability region, and conjugative transfer region. However, IncP plasmids are the most heterogeneous, and they are not identical to one another. The replication region of IncP plasmids mainly consists of two gene loci: oriV (origin of replication) and trfA (encoding the protein necessary for oriV activation). Genes related to the maintenance region include the par region (parA, parB, parC, parD, and parE), korA-kfrA region, and Tn1 (present in IncPα plasmids and absent in IncPβ plasmids). Some plasmid backbones contain the higA-B toxin-antitoxin system. The conjugative transfer region consists of the Tra1 and Tra2 regions. The backbone genes of IncPα and IncPβ plasmids mainly differ in the region between oriV and trfA, as well as between Tra1 and Tra2 ( Pansegrau et al., 1994; Zhao et al., 2017). Antibiotic resistance genes and mobility elements are also present in the accessory modules of IncP plasmids.
Among the β-lactam antibiotic resistance genes carried by IncP plasmids, the bla KPC gene has the highest carriage rate, followed by bla OXA, bla GES, bla IMP, and mcr. The most common subtypes of bla KPC resistance gene are bla KPC-2, and bla OXA-2 subtype also commonly appears ( Figure 3 ). On IncP plasmids, additional genes for β-lactam antibiotic resistance have also been identified, including bla TEM, bla VIM, bla SHV, and bla NPS. Additionally, frequent co-occurrence of bla OXA-48 and bla IMP-45 has been observed on IncP plasmids ( Table 2 ), potentially forming stable gene cassette sequences (Zhang et al., 2021). Due to the complex gene structure of IncP plasmids, the structure of the carried resistance genes is also quite disordered. No conserved sequences have been found, but some plasmids’ resistance genes are often located downstream of Class I integrons or transposons such as Tn3, Tn402, or their derivatives. This may indicate the presence of integration hotspots associated with Class I integrons and/or transposons Tn402, facilitating the dissemination of resistance genes. IncP plasmids harbor other resistance genes, including aacA, aadA, aacC, tetAR, mer, sul1, qacEΔ1, strAB, dhfrllc, aac (6’)-lb, aac (2’)-IIa, and fosA. They also demonstrate resistance to heavy metals such as Ni, Cu, Zn, and Pb (Yoshii et al., 2015).
Similar to the IncHI plasmid, The IncP plasmid’s conjugation transfer is regulated by the Tra1 and Tra2 regions. The Tra1 region contains core genes such as traF, traG, traH, traI, traJ, traK, traL, and traM, which are responsible for DNA transfer and replication. The Tra2 region consists of trbB, trbC, trbD, trbE, trbF, trbG, trbH, trbI, trbJ, trbK, and trbL, and its main purpose is to facilitate contact between donor and recipient bacteria, as well as the formation of Mpf. TrbB and TrbE are involved in the pili assembly process, while traI, traH, traJ, and traK are involved in DNA processing. TraM can increase the frequency of conjugation transfer. Although most IncP plasmids possess conjugation transfer genes, the transfer frequencies vary significantly. It is not possible to analyze the reasons for this difference through the detection of gene sequence variations. This may be attributed to the incompleteness or specificity of the conjugation transfer genes in IncP plasmids (Yano et al., 2013).
Unlike other plasmids, the IncP plasmid has host bacteria that include Pseudomonas species, indicating an even broader host range. The IncP plasmid often carries the bla KPC-2 or bla OXA-2 genes. Additionally, there have been reports of more structurally complex IncP megaplasmids. These characteristics present significant challenges for clinical anti-infective treatment.
6. Conclusions
Incompatible plasmids have appeared in bacteria previously unexplored, along with the continuous enrichment and dissemination of the harbored antibiotic resistance genes, adding a new level to the spread of bacterial resistance genes and becoming the main platform for the transmission of antibiotic resistance genes. The geographical distribution of plasmids also affects the spread of antibiotic-resistant genes. There are certain differences in bacterial populations in different geographical regions, which may be related to human activities, environmental factors, and the use of antibiotics. Each type of plasmid possesses a basic and highly conserved plasmid backbone. Additionally, the high variability of accessory modules carried by plasmids allows for greater diversity among different types of plasmids. These accessory modules contain various genetic elements, which not only carry multiple gene cassettes and resistance genes but also have the ability to capture exogenous genes or provide promoters for resistance genes. The presence of genetic elements provides convenient conditions for the capture, dissemination, and expression of resistance genes, further increasing the rate of resistance formation and dissemination. Different types of incompatible plasmids carry different β-lactamase genes, while resistance genes for sulfonamides and aminoglycosides are also common. With further research, an increasing number of resistance genes associated with plasmids are being discovered. This urgently requires us to conduct more in-depth studies on different genetic elements to establish a comprehensive knowledge system for the enrichment and dissemination of resistance genes, in order to effectively control the spread of multi-drug-resistant bacteria.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a grant from the Henan Provincial Department of Science and Technology Research Project (No. 232102310176).
Author contributions
LF: Data curation, Writing – original draft. RC: Writing – review & editing. CL: Data curation, Software, Writing – original draft. JS: Formal analysis, Methodology, Writing – original draft. RL: Data curation, Software, Writing – original draft. YS: Data curation, Writing – original draft. XG: Conceptualization, Funding acquisition, Methodology, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1472876/full#supplementary-material
References
- Adamczuk M., Zaleski P., Dziewit L., Wolinowska R., Nieckarz M., Wawrzyniak P., et al. (2015). Diversity and global distribution of incL/M plasmids enabling horizontal dissemination of β-lactam resistance genes among the Enterobacteriaceae. BioMed. Res. Int. 2015, 414681. doi: 10.1155/2015/414681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk M., Jagura-Burdzy G. (2003). Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50, 425–453. doi: 10.18388/abp.2003_3696 [DOI] [PubMed] [Google Scholar]
- Al Mamun A. A. M., Kishida K., Christie P. J. (2021). Protein transfer through an F plasmid-encoded type IV secretion system suppresses the mating-induced SOS response. mBio 12, e0162921. doi: 10.1128/mBio.01629-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose S. J., Harmer C. J., Hall R. M. (2018). Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid 96-97, 7–12. doi: 10.1016/j.plasmid.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Ammar A. M., Abd El-Aziz N. K., Aggour M. G., Ahmad A. A. M., Abdelkhalek A., Muselin F., et al. (2024). A newly incompatibility F replicon allele (FIB81) in extensively drug-resistant Escherichia coli isolated from diseased broilers. Int. J. Mol. Sci. 25 (15) (8347), 8437. doi: 10.3390/ijms25158347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. A., Mir R. A., Qadri H., Dhiman R., Almilaibary A., Alkhanani M., et al. (2023). Integrons in the development of antimicrobial resistance: critical review and perspectives. Front. Microbiol. 14, 1231938. doi: 10.3389/fmicb.2023.1231938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J. M., Webber M. A., Baylay A. J., Ogbolu D. O., Piddock L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380 [DOI] [PubMed] [Google Scholar]
- Bragagnolo N., Rodriguez C., Samari-Kermani N., Fours A., Korouzhdehi M., Lysenko R., et al. (2020). Protein dynamics in F-like bacterial conjugation. Biomedicines 8 (9), 362. doi: 10.3390/biomedicines8090362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Seiffert S. N., Schwendener S., Perreten V., Endimiani A. (2015). Differentiation of incL and incM plasmids associated with the spread of clinically relevant antimicrobial resistance. PloS One 10, e0123063. doi: 10.1371/journal.pone.0123063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Villa L., Poirel L., Bonnin R. A., Nordmann P. (2012). Evolution of IncA/C blaCMY-2-carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 56, 783–786. doi: 10.1128/AAC.05116-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N., Sauvé M., Matteau D., Lauzon G., Rodrigue S., Burrus V. (2014). Development of pVCR94ΔX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front. Microbiol. 5, 44. doi: 10.3389/fmicb.2014.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabbert Y. A., Scavizzi M. R., Witchitz J. L., Gerbaud G. R., Bouanchaud D. H. (1972). Incompatibility groups and the classification of fi - resistance factors. J. Bacteriol. 112, 666–675. doi: 10.1128/jb.112.2.666-675.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaslus-Dancla E., Lafont J. P. (1985). IncH plasmids in Escherichia coli strains isolated from broiler chicken carcasses. Appl. Environ. Microbiol. 49, 1016–1018. doi: 10.1128/aem.49.4.1016-1018.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., He L., Li Y., Zeng Z., Deng Y., Liu Y., et al. (2014). Complete sequence of a F2:A-:B- plasmid pHN3A11 carrying rmtB and qepA, and its dissemination in China. Vet. Microbiol. 174, 267–271. doi: 10.1016/j.vetmic.2014.08.023 [DOI] [PubMed] [Google Scholar]
- Chen Y. T., Lauderdale T. L., Liao T. L., Shiau Y. R., Shu H. Y., Wu K. M., et al. (2007). Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 beta-lactamases in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 51, 3004–3007. doi: 10.1128/AAC.00167-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Li C., Ge H., Qiao J., Fang L., Liu C., et al. (2024). Difference analysis and characteristics of incompatibility group plasmid replicons in gram-negative bacteria with different antimicrobial phenotypes in Henan, China. BMC Microbiol. 24, 64. doi: 10.1186/s12866-024-03212-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. T., Liao T. L., Liu Y. M., Lauderdale T. L., Yan J. J., Tsai S. F. (2009). Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob. Agents Chemother. 53, 1235–1237. doi: 10.1128/AAC.00970-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte D., Mesa D., Jové T., Zamparette C. P., Sincero T. C. M., Palmeiro J. K., et al. (2022). Novel Insights into bla(GES) Mobilome Reveal Extensive Genetic Variation in Hospital Effluents. Microbiol. Spectr. 10, e0246921. doi: 10.1128/spectrum.02469-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Castillo C. S., Hikima J., Jang H. B., Nho S. W., Jung T. S., Wongtavatchai J., et al. (2013). Comparative sequence analysis of a multidrug-resistant plasmid from Aeromonas hydrophila. Antimicrob. Agents Chemother. 57, 120–129. doi: 10.1128/AAC.01239-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo C. S., Jang H. B., Hikima J., Jung T. S., Morii H., Hirono I., et al. (2013). Comparative analysis and distribution of pP9014, a novel drug resistance IncP-1 plasmid from Photobacterium damselae subsp. piscicida. Int. J. Antimicrob. Agents. 42, 10–18. doi: 10.1016/j.ijantimicag.2013.02.027 [DOI] [PubMed] [Google Scholar]
- Dhanji H., Murphy N. M., Akhigbe C., Doumith M., Hope R., Livermore D. M., et al. (2011). Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum β-lactamase from UK river water. J. Antimicrob. Chemother. 66, 512–516. doi: 10.1093/jac/dkq472 [DOI] [PubMed] [Google Scholar]
- Dias J. B., Soncini J. G. M., Cerdeira L., Lincopan N., Vespero E. C. (2022). MDR Escherichia coli carrying CTX-M-24 (IncF[F-:A1:B32]) and KPC-2 (IncX3/IncU) plasmids isolated from community-acquired urinary trainfection in Brazil. Braz. J. Infect. Dis. 26, 102706. doi: 10.1016/j.bjid.2022.102706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pilato V., Pollini S., Miriagou V., Rossolini G. M., D’Andrea M. M. (2024). Carbapenem-resistant Klebsiella pneumoniae: the role of plasmids in emergence, dissemination, and evolution of a major clinical challenge. Expert Rev. Anti Infect. Ther. 22, 25–43. doi: 10.1080/14787210.2024.2305854 [DOI] [PubMed] [Google Scholar]
- Doublet B., Boyd D., Douard G., Praud K., Cloeckaert A., Mulvey M. R. (2012). Complete nucleotide sequence of the multidrug resistance IncA/C plasmid pR55 from Klebsiella pneumoniae isolated in 1969. J. Antimicrob. Chemother. 67, 2354–2360. doi: 10.1093/jac/dks251 [DOI] [PubMed] [Google Scholar]
- Fricke W. F., Welch T. J., McDermott P. F., Mammel M. K., LeClerc J. E., White D. G., et al. (2009). Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191, 4750–4757. doi: 10.1128/JB.00189-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Koraimann G. (2010). Regulation of bacterial conjugation: balancing opportunity with adversity. Future Microbiol. 5, 1057–1071. doi: 10.2217/fmb.10.70 [DOI] [PubMed] [Google Scholar]
- Fukuda A., Nakano H., Suzuki Y., Nakajima C., Usui M. (2022). Conjugative IncHI2/HI2A plasmids harbouring mcr-9 in colistin-susceptible Escherichia coli isolated from diseased pigs in Japan. Access Microbiol. 4, acmi000454. doi: 10.1099/acmi.0.000454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galata V., Fehlmann T., Backes C., Keller A. (2019). PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–d202. doi: 10.1093/nar/gky1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancz A., Kondratyeva K., Cohen-Eli D., Navon-Venezia S. (2021). Genomics and virulence of klebsiella pneumoniae kpnu95 ST1412 harboring a novel incf plasmid encoding blactx-M-15 and qnrs1 causing community urinary tract infection. Microorganisms 9 (5), 1022. doi: 10.3390/microorganisms9051022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Fernández A., Miriagou V., Papagiannitsis C. C., Giordano A., Venditti M., Mancini C., et al. (2010). An ertapenem-resistant extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob. Agents Chemother. 54, 4178–4184. doi: 10.1128/AAC.01301-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcillán-Barcia M. P., Redondo-Salvo S., de la Cruz F. (2023). Plasmid classifications. Plasmid 126, 102684. doi: 10.1016/j.plasmid.2023.102684 [DOI] [PubMed] [Google Scholar]
- Geoffroy F., Uecker H. (2023). Limits to evolutionary rescue by conjugative plasmids. Theor. Popul. Biol. 154, 102–117. doi: 10.1016/j.tpb.2023.10.001 [DOI] [PubMed] [Google Scholar]
- Ghaly T. M., Tetu S. G., Penesyan A., Qi Q., Rajabal V., Gillings M. R. (2022). Discovery of integrons in Archaea: Platforms for cross-domain gene transfer. Sci. Adv. 8, eabq6376. doi: 10.1126/sciadv.abq6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour M. W., Thomson N. R., Sanders M., Parkhill J., Taylor D. E. (2004). The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52, 182–202. doi: 10.1016/j.plasmid.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Gordils-Valentin L., Ouyang H., Qian L., Hong J., Zhu X. (2024). Conjugative type IV secretion systems enable bacterial antagonism that operates independently of plasmid transfer. Commun. Biol. 7, 499. doi: 10.1038/s42003-024-06192-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lynne A. M., David D. E., Tang H., Xu J., Nayak R., et al. (2012). DNA sequence analysis of plasmids from multidrug resistant Salmonella enterica serotype Heidelberg isolates. PloS One 7, e51160. doi: 10.1371/journal.pone.0051160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer C. J., Hall R. M. (2015). The A to Z of A/C plasmids. Plasmid 80, 63–82. doi: 10.1016/j.plasmid.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Harmer C. J., Hamidian M., Hall R. M. (2017). pIP40a, a type 1 IncC plasmid from 1969 carries the integrative element GIsul2 and a novel class II mercury resistance transposon. Plasmid 92, 17–25. doi: 10.1016/j.plasmid.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Hayakawa M., Tokuda M., Kaneko K., Nakamichi K., Yamamoto Y., Kamijo T., et al. (2022). Hitherto-unnoticed self-transmissible plasmids widely distributed among different environments in Japan. Appl. Environ. Microbiol. 88, e0111422. doi: 10.1128/aem.01114-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Kontomichalou P., Smith J. T. (1974). Molecular specificities of R factor-determined beta-lactamases: correlation with plasmid compatibility. J. Bacteriol. 117, 56–62. doi: 10.1128/jb.117.1.56-62.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hounmanou Y. M. G., Bortolaia V., Dang S. T. T., Truong D., Olsen J. E., Dalsgaard A. (2021). ESBL and ampC β-lactamase encoding genes in E. coli from pig and pig farm workers in Vietnam and their association with mobile genetic elements. Front. Microbiol. 12, 629139. doi: 10.3389/fmicb.2021.629139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Cao M., Hu Y., Zhang R., Xiao Y., Chen G. (2021). Prevalence and mechanisms of fosfomycin resistance among KPC-producing Klebsiella pneumoniae clinical isolates in China. Int. J. Antimicrob. Agents. 57, 106226. doi: 10.1016/j.ijantimicag.2020.106226 [DOI] [PubMed] [Google Scholar]
- Johnson T. J., Lang K. S. (2012). IncA/C plasmids: An emerging threat to human and animal health? Mob. Genet. Elements 2, 55–58. doi: 10.4161/mge.19626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. J., Wannemeuhler Y. M., Scaccianoce J. A., Johnson S. J., Nolan L. K. (2006). Complete DNA sequence, comparative genomics, and prevalence of an IncHI2 plasmid occurring among extraintestinal pathogenic Escherichia coli isolates. Antimicrob. Agents Chemother. 50, 3929–3933. doi: 10.1128/AAC.00569-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karampatakis T., Tsergouli K., Behzadi P. (2023). Carbapenem-resistant Klebsiella pneumoniae: virulence factors, molecular epidemiology and latest updates in treatment options. Antibiotics (Basel). 12 (2), 234. doi: 10.3390/antibiotics12020234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J., Hirono I., Kurokawa K., Maki T., Hawke J., Kondo H., et al. (2008). Complete DNA sequence and analysis of the transferable multiple-drug resistance plasmids (R Plasmids) from Photobacterium damselae subsp. piscicida isolates collected in Japan and the United States. Antimicrob. Agents Chemother. 52, 606–611. doi: 10.1128/AAC.01216-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T. D., Gilmour M. W., Gunton J. E., Standeven L. J., Taylor D. E. (2002). Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 184, 2173–2180. doi: 10.1128/JB.184.8.2173-2180.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley T. D., Gilmour M. W., Gunton J. E., Tracz D. M., Taylor D. E. (2003). Functional and mutational analysis of conjugative transfer region 2 (Tra2) from the IncHI1 plasmid R27. J. Bacteriol. 185, 581–591. doi: 10.1128/JB.185.2.581-591.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J. (1952). Cell genetics and hereditary symbiosis. Physiol. Rev. 32, 403–430. doi: 10.1152/physrev.1952.32.4.403 [DOI] [PubMed] [Google Scholar]
- Liang H., Li X., Yan H. (2022). Identification of a novel incHI1B plasmid in MDR Klebsiella pneumoniae 200 from swine in China. Antibiotics (Basel). 11 (9), 1225. doi: 10.3390/antibiotics11091225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Yin Z., Zhao Y., Liang L., Feng J., Zhan Z., et al. (2017). Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int. J. Antimicrob. Agents. 49, 709–718. doi: 10.1016/j.ijantimicag.2017.01.021 [DOI] [PubMed] [Google Scholar]
- Lowbury E. J., Lilly H. A., Kidson A., Ayliffe G. A., Jones R. J. (1969). Sensitivity of Pseudomonas aeruginosa to antibiotics: emergence of strains highly resistant to carbenicillin. Lancet 2, 448–452. doi: 10.1016/S0140-6736(69)90163-9 [DOI] [PubMed] [Google Scholar]
- Luo Q., Lu P., Chen Y., Shen P., Zheng B., Ji J., et al. (2024). ESKAPE in China: epidemiology and characteristics of antibiotic resistance. Emerg. Microbes Infect. 13, 2317915. doi: 10.1080/22221751.2024.2317915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Yu W., Zhou K., Guo L., Shen P., Lu H., et al. (2017). Molecular Epidemiology and Colistin Resistant Mechanism of mcr-Positive and mcr-Negative Clinical Isolated Escherichia coli . Front. Microbiol. 8, 2262. doi: 10.3389/fmicb.2017.02262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macesic N., Hawkey J., Vezina B., Wisniewski J. A., Cottingham H., Blakeway L. V., et al. (2023). Genomic dissection of endemic carbapenem resistance reveals metallo-beta-lactamase dissemination through clonal, plasmid and integron transfer. Nat. Commun. 14, 4764. doi: 10.1038/s41467-023-39915-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J., Chandler M. (1998). Insertion sequences. Microbiol. Mol. Biol. Rev. 62, 725–774. doi: 10.1128/MMBR.62.3.725-774.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y., Yamamoto M., Nagao M., Ito Y., Takakura S., Ichiyama S. (2013). Association of fluoroquinolone resistance, virulence genes, and IncF plasmids with extended-spectrum-β-lactamase-producing Escherichia coli sequence type 131 (ST131) and ST405 clonal groups. Antimicrob. Agents Chemother. 57, 4736–4742. doi: 10.1128/AAC.00641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzejewska J., Kulińska A., Jagura-Burdzy G. (2007). Functional analysis of replication and stability regions of broad-host-range conjugative plasmid CTX-M3 from the IncL/M incompatibility group. Plasmid 57, 95–107. doi: 10.1016/j.plasmid.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Mori N., Tada T., Oshiro S., Kuwahara-Arai K., Kirikae T., Uehara Y. (2021). A transferrable IncL/M plasmid harboring a gene encoding IMP-1 metallo-β-lactamase in clinical isolates of Enterobacteriaceae. BMC Infect. Dis. 21, 1061. doi: 10.1186/s12879-021-06758-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluwadare M., Lee M. D., Grim C. J., Lipp E. K., Cheng Y., Maurer J. J. (2020). The Role of the Salmonella spvB IncF Plasmid and Its Resident Entry Exclusion Gene traS on Plasmid Exclusion. Front. Microbiol. 11, 949. doi: 10.3389/fmicb.2020.00949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei Sekyere J., Maningi N. E., Modipane L., Mbelle N. M. (2020). Emergence of mcr-9.1 in extended-spectrum-β-lactamase-producing clinical Enterobacteriaceae in Pretoria, South Africa: global evolutionary phylogenomics, resistome, and mobilome. mSystems 5 (3), e00148-20. doi: 10.1128/mSystems.00148-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota Y., Prah I., Nukui Y., Koike R., Saito R. (2022). bla(KPC-2)-Encoding IncP-6 Plasmids in Citrobacter freundii and Klebsiella variicola Strains from Hospital Sewage in Japan. Appl. Environ. Microbiol. 88, e0001922. doi: 10.1128/aem.00019-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E., Barth P. T., Figurski D. H., Guiney D. G., Haas D., et al. (1994). Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239, 623–663. doi: 10.1006/jmbi.1994.1404 [DOI] [PubMed] [Google Scholar]
- Park Y. J., Kim S. Y., Yu J. K., Kim S. I., Uh Y., Hong S. G., et al. (2009). Spread of Serratia marcescens coharboring aac(6’)-Ib-cr, bla CTX-M, armA, and bla OXA-1 carried by conjugative IncL/M type plasmid in Korean hospitals. Microb. Drug Resist. 15, 97–102. doi: 10.1089/mdr.2009.0867 [DOI] [PubMed] [Google Scholar]
- Parkhill J., Dougan G., James K. D., Thomson N. R., Pickard D., Wain J., et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852. doi: 10.1038/35101607 [DOI] [PubMed] [Google Scholar]
- Partridge S. R., SM K., Firth N., Jensen S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31 (4), e00088-17. doi: 10.1128/CMR.00088-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Périchon B., Bogaerts P., Lambert T., Frangeul L., Courvalin P., Galimand M. (2008). Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob. Agents Chemother. 52, 2581–2592. doi: 10.1128/AAC.01540-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popowska M., Krawczyk-Balska A. (2013). Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4, 44. doi: 10.3389/fmicb.2013.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potron A., Poirel L., Nordmann P. (2014). Derepressed transfer properties leading to the efficient spread of the plasmid encoding carbapenemase OXA-48. Antimicrob. Agents Chemother. 58, 467–471. doi: 10.1128/AAC.01344-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada A. M., Correa A., Restrepo E., Capataz C. (2022). Escherichia coli ST471 producing VIM-4 metallo-β-lactamase in Colombia. Microb. Drug Resist. 28, 288–292. doi: 10.1089/mdr.2021.0031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozwandowicz M., Kant A., Wagenaar J., Mevius D., Hordijk J., Brouwer M. (2023). Understanding the genetic basis of the incompatibility of IncK1 and IncK2 plasmids. Open Res. Eur. 3, 53. doi: 10.12688/openreseurope [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra S. Y., Diaz L., Wiesner M., Correa A., Arévalo S. A., Reyes J., et al. (2017). Genomic and molecular characterization of clinical isolates of Enterobacteriaceae harboring mcr-1 in Colombia, 2002 to 2016. Antimicrob. Agents Chemother. 61 (12), e00841-17. doi: 10.1128/AAC.00841-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter A., Heuer H., Szczepanowski R., Poler S. M., Schneiker S., Pühler A., et al. (2005). Plasmid pB8 is closely related to the prototype IncP-1beta plasmid R751 but transfers poorly to Escherichia coli and carries a new transposon encoding a small multidrug resistance efflux protein. Plasmid 54, 135–148. doi: 10.1016/j.plasmid.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Schlüter A., Nordmann P., Bonnin R. A., Millemann Y., Eikmeyer F. G., Wibberg D., et al. (2014). IncH-type plasmid harboring bla CTX-M-15, bla DHA-1, and qnrB4 genes recovered from animal isolates. Antimicrob. Agents Chemother. 58, 3768–3773. doi: 10.1128/AAC.02695-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmartz G. P., Hartung A., Hirsch P., Kern F., Fehlmann T., Müller R., et al. (2022). PLSDB: advancing a comprehensive database of bacterial plasmids. Nucleic Acids Res. 50, D273–D2d8. doi: 10.1093/nar/gkab1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherburne C. K., Lawley T. D., Gilmour M. W., Blattner F. R., Burland V., Grotbeck E., et al. (2000). The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28, 2177–2186. doi: 10.1093/nar/28.10.2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P., Gourbeyre E., Chandler M. (2014). Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 38, 865–891. doi: 10.1111/1574-6976.12067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C., Arismendi T., Wright M., Hartman A., Gonzalez A., Gill M., et al. (2020). F plasmids are the major carriers of antibiotic resistance genes in human-associated commensal Escherichia coli . mSphere 5 (4), e00709-20. doi: 10.1128/msphere.00709-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino Y., Hirota Y. (1962). Conjugal fertility associated with resistance factor R in Escherichia coli . J. Bacteriol. 84, 902–910. doi: 10.1128/jb.84.5.902-910.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. E. (2009). Thermosensitive nature of IncHI1 plasmid transfer. Antimicrob. Agents Chemother. 53, 2703. doi: 10.1128/AAC.00230-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin L. A., Jarocki V. M., Kenyon J., Drigo B., Donner E., Djordjevic S. P., et al. (2024). Genomic analysis of diverse environmental Acinetobacter isolates identifies plasmids, antibiotic resistance genes, and capsular polysaccharides shared with clinical strains. Appl. Environ. Microbiol. 90, e0165423. doi: 10.1128/aem.01654-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton J. F., Pike R., Perry C., Jenkins C., Turton J. A., Meunier D., et al. (2022). Wide distribution of Escherichia coli carrying IncF plasmids containing bla (NDM-5) and rmtB resistance genes from hospitalized patients in England. J. Med. Microbiol. 71 (8), 10.1099/jmm.0.001569. doi: 10.1099/jmm.0.001569 [DOI] [PubMed] [Google Scholar]
- Vázquez X., García P., García V., de Toro M., Ladero V., Heinisch J. J., et al. (2021). Genomic analysis and phylogenetic position of the complex IncC plasmid found in the Spanish monophasic clone of Salmonella enterica serovar Typhimurium. Sci. Rep. 11, 11482. doi: 10.1038/s41598-021-90299-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa L., Carattoli A. (2020). Plasmid typing and classification. Methods Mol. Biol. 2075, 309–321. doi: 10.1007/978-1-4939-9877-7_22 [DOI] [PubMed] [Google Scholar]
- Wang Q., Wang W., Zhu Q., Shoaib M., Chengye W., Zhu Z., et al. (2024). The prevalent dynamic and genetic characterization of mcr-1 encoding multidrug resistant Escherichia coli strains recovered from poultry in Hebei, China. J. Glob. Antimicrob. Resist. 38, 354–362. doi: 10.1016/j.jgar.2024.04.001 [DOI] [PubMed] [Google Scholar]
- Wu S., Feng Y., Yang Y., Zhu K., Wang S., Wang X., et al. (2023). Characterization of a novel carbapenem-hydrolysing β-lactamase OXA-1041 in Escherichia coli . J. Antimicrob. Chemother. 78, 1288–1294. doi: 10.1093/jac/dkad091 [DOI] [PubMed] [Google Scholar]
- Yakobson E., Guiney G. (1983). Homology in the transfer origins of broad host range IncP plasmids: definition of two subgroups of P plasmids. Mol. Gen. Genet. 192, 436–438. doi: 10.1007/BF00392187 [DOI] [PubMed] [Google Scholar]
- Yang W., Hu F. (2022). Research updates of plasmid-mediated aminoglycoside resistance 16S rRNA methyltransferase. Antibiotics (Basel). 11 (7), 906. doi: 10.3390/antibiotics11070906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H., Rogers L. M., Knox M. G., Heuer H., Smalla K., Brown C. J., et al. (2013). Host range diversification within the IncP-1 plasmid group. Microbiol. (Reading) 159, 2303–2315. doi: 10.1099/mic.0.068387-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii A., Omatsu T., Katayama Y., Koyama S., Mizutani T., Moriyama H., et al. (2015). Two types of genetic carrier, the IncP genomic island and the novel IncP-1β plasmid, for the aac(2’)-IIa gene that confers kasugamycin resistance in Acidovorax avenae ssp. avenae. Mol. Plant Pathol. 16, 288–300. doi: 10.1111/mpp.2015.16.issue-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang L., Li D., Li P., Yuan L., Yang F., et al. (2021). An IncP-2 plasmid sublineage associated with dissemination of bla(IMP-45) among carbapenem-resistant Pseudomonas aeruginosa. Emerg. Microbes Infect. 10, 442–449. doi: 10.1080/22221751.2021.1894903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Xie Y., Zhang Y., Lei T., Zhou L., Yao J., et al. (2024). Evolution of ceftazidime-avibactam resistance driven by mutations in double-copy bla(KPC-2) to bla(KPC-189) during treatment of ST11 carbapenem-resistant Klebsiella pneumoniae. mSystems 9, e0072224. doi: 10.1128/msystems.00722-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Ye X., Yin Z., Hu M., Wang B., Liu W., et al. (2022). Comparative genomics reveals new insights into the evolution of the IncA and IncC family of plasmids. Front. Microbiol. 13, 1045314. doi: 10.3389/fmicb.2022.1045314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Feng Y., Lü X., McNally A., Zong Z. (2017). IncP Plasmid Carrying Colistin Resistance Gene mcr-1 in Klebsiella pneumoniae from Hospital Sewage. Antimicrob. Agents Chemother. 61 (2), e02229-16. doi: 10.1128/AAC.02229-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. Y., Zhu Y. Q., Li Y. N., Mu X. D., You L. P., Xu C., et al. (2015). Coexistence of SFO-1 and NDM-1 β-lactamase genes and fosfomycin resistance gene fosA3 in an Escherichia coli clinical isolate. FEMS Microbiol. Lett. 362, 1–7. doi: 10.1093/femsle/fnu018 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Ye L., Chan E. W., Chen S. (2019). Identification and characterization of a conjugative blaVIM-1-bearing plasmid in Vibrio alginolyticus of food origin. J. Antimicrob. Chemother. 74, 1842–1847. doi: 10.1093/jac/dkz140 [DOI] [PubMed] [Google Scholar]
- Zhu W. H., Luo L., Wang J. Y., Zhuang X. H., Zhong L., Liao K., et al. (2009). Complete nucleotide sequence of pCTX-M360, an intermediate plasmid between pEL60 and pCTX-M3, from a multidrug-resistant Klebsiella pneumoniae strain isolated in China. Antimicrob. Agents Chemother. 53, 5291–5293. doi: 10.1128/AAC.00032-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.