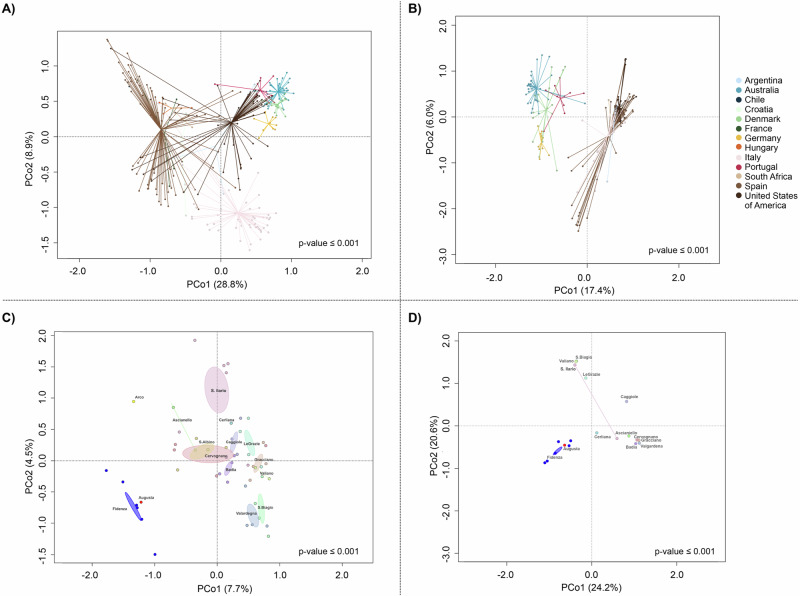
Fig. 2. The bulk soil microbiome of Montepulciano vineyards shows a clear differentiation compared to other vineyards around the world.
Comparisons were made for both 16S rRNA and ITS sequencing, using data from Gobbi et al.8. A Principal coordinate analysis (PCoA) based on unweighted UniFrac distances showing the variation of Vitis vinifera cultivar Sangiovese bulk soil bacterial composition at a wide geographical scale (worldwide), including Montepulciano samples in the Italian site (permutation test with pseudo-F ratio, p-value ≤ 0.001). B PCoA based on Bray–Curtis distances showing the variation of V. vinifera cultivar Sangiovese bulk soil fungal composition at a wide geographical scale (worldwide), including Montepulciano samples in the Italian site (p-value ≤ 0.001). C The same graph as in (A), but at a finer geographical scale, including only Italian samples from Gobbi et al.8 and Montepulciano samples (p-value ≤ 0.001). D The same graph as in (B), but at a finer geographical scale, including only Italian samples from Gobbi et al.8 and Montepulciano samples (p-value ≤ 0.001). For C and D, sample origin is indicated on each graph. For all PCoA plots, the first and second principal components are plotted and the percentage of variance in the dataset explained by each axis is shown.