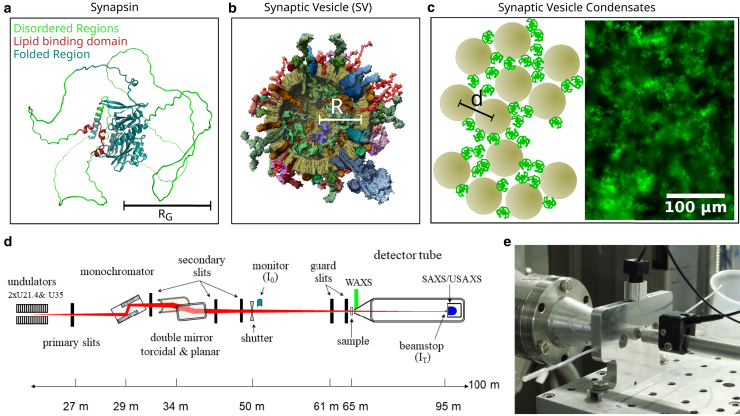
Figure 1.
(a) Structure of synapsin as predicted by AlphaFold (16). The disordered regions and lipid binding domain are indicated in green and red, respectively. (b) Schematic structure of SV as shown in (2). The vesicle radius R and the density profile including the bilayer and the protein shells can be determined from the SV form factor measured by solution scattering. (c, left) Schematic structure of a synapsin vesicle condensate, with the average distance d between neighboring vesicles, which can be inferred from the structure factor. (c, right) Fluorescence microscopy image (20×) of condensates formed from EGFP-labeled synapsin and lipid vesicles (), adapted from (11). While a fractal appearance is readily visible on large scales, the present work addresses the condensate morphology on sub-μm scales by way of SAXS. (d) Schematic layout and optical components of the ID02 beamline at ESRF as shown in (17). The SAXS data was recorded at two different sample-detector distances of and , respectively. (e) Photograph of the beamline near the sample, showing the sample chamber including the flowthrough capillary.