Fig. 4:
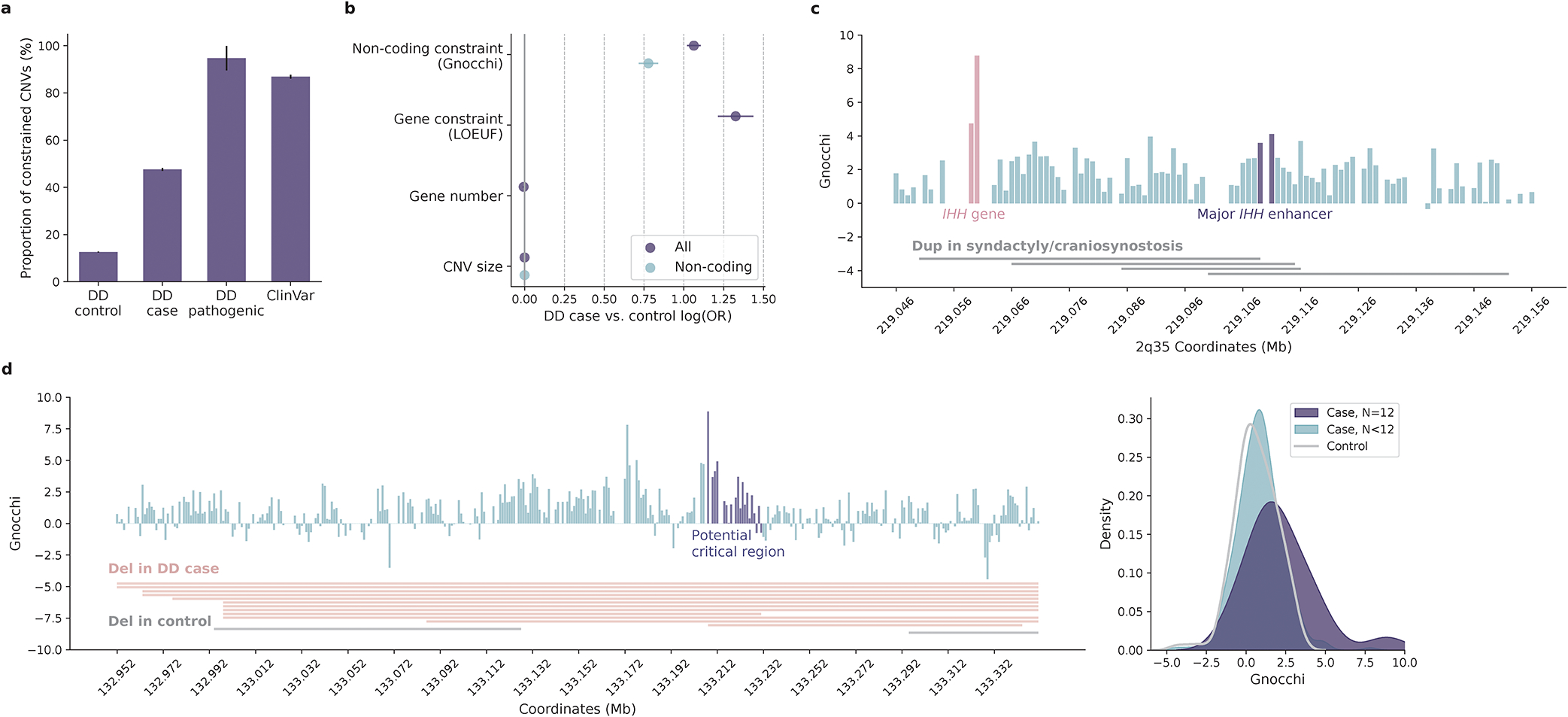
Contribution of non-coding constraint in evaluating copy number variants (CNVs). a, Proportions of constrained CNVs (Gnocchi≥4) identified in individuals with developmental delay (DD cases) versus healthy controls. Constrained CNVs are more common in DD cases than controls (7,239/17,004=42.6% versus 10,403/83,526=12.5%) and are most frequent for CNVs previously implicated as pathogenic (18/19=94.7% by DD and 3,433/4,014=85.5% by ClinVar). Error bars indicate standard errors of the proportions. b, Contribution of non-coding constraint to predicting CNVs in DD cases versus controls. Non-coding constraint remains a significant predictor for the case/control status of CNVs after adjusting for gene constraint (LOEUF score), gene number, and size of CNVs (Ncase=17,004, Ncontrol=83,526; purple), as well as being tested in the subset of non-coding CNVs (Ncase=8,702, Ncontrol=66,795; blue). Error bars indicate 95% confidence intervals of the log odds ratios. c, CNVs at the IHH locus associated with synpolydactyly and craniosynostosis. The four implicated duplications (grey horizontal bars) span a ~102kb sequence upstream of IHH. Each vertical bar shows the Gnocchi score of a 1kb window within the locus, with the highest score overlapping the IHH gene (red) and the highest non-coding score overlapping the major IHH enhancers (purple); gaps indicate windows removed by quality filters. d, Non-coding CNVs with the highest Gnocchi score identified in DD cases. The highest-scored window is located within the potential “critical region” (purple vertical bars) shared by 12 DD deletions (red horizontal bars; grey indicates two deletions observed in controls). The critical region overall, has a significantly higher Gnocchi score than the other regions affected by DD or control deletions, as shown in the kernel density estimate (KDE) plot on the right.
