Figure 2.
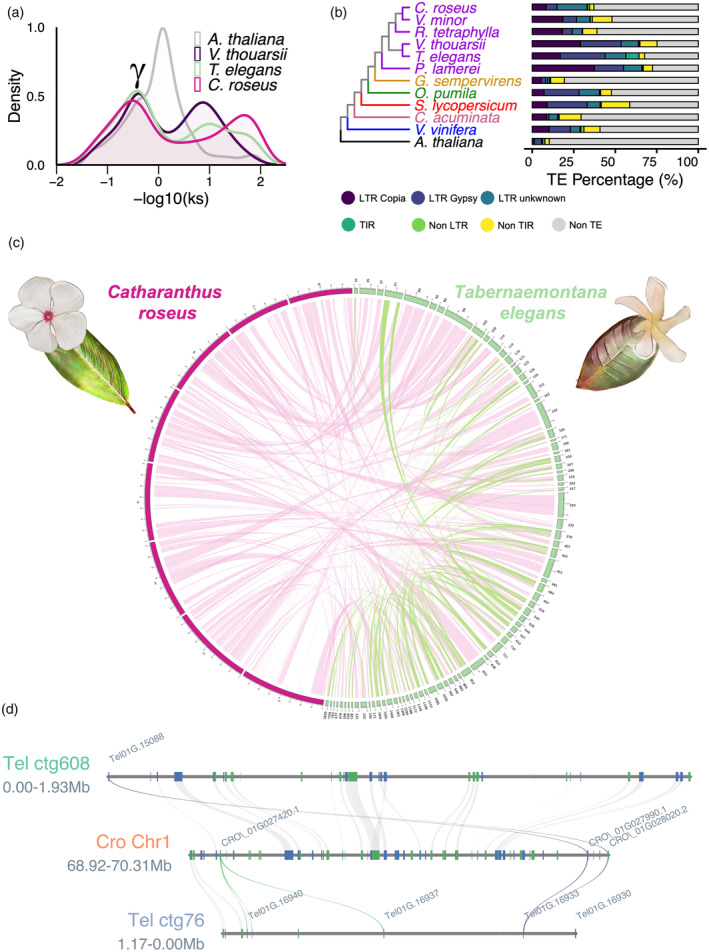
The annotated Tabernaemontana elegans genome.
(a) Synonymous substitution rate (Ks) distribution plot for T. elegans paralogs compared to Arabidopsis thaliana, Voacanga thouarsii, and Catharanthus roseus. γ indicates the conserved γ whole‐genome triplication event that is shared among Eudicotyledons.
(b) Phylogenetic tree of T. elegans and 11 other species, including five Apocynaceae (purple: C. roseus, Vinca minor, Rauvolfia tetraphylla, V. thouarsii, and Pachypodium lamerei), one Gelsemiaceae (yellow: Gelsemium sempervirens), one Rubiaceae (green: Ophiorrhiza pumila), one Solanaceae (red: Solanum lycopersicum), one Nyssaceae (pink: Camptotheca acuminata), one Vitaceae (blue: Vitis vinifera), and one Brassicaceae (black: A. thaliana).
(c) Circos plot highlighting T. elegans genome‐wide self‐synteny (green links) and synteny between the T. elegans and C. roseus genomes (pink links). The eight C. roseus chromosomes are displayed in pink. Only the 80 T. elegans contigs containing 80 or more genes are shown in green.
(d) Focus on the T. elegans genomic regions encompassing the PYS gene and the corresponding syntenic region retrieved in C. roseus. CYP71 (purple, PYS; green, other CYP71s) and gene synteny blocks are indicated by colored and gray lines, respectively.
