Figure 6.
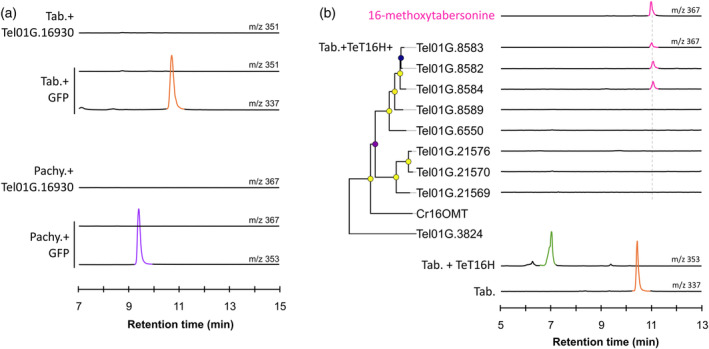
Screening of methyltransferase activities for methylation of tabersonine derivatives.
(a) Screening of N‐methyltransferase activity with tabersonine and pachysiphine. Positive differential mass chromatograms of tabersonine at m/z 337, putative N‐methyltabersonine at m/z 351, pachysiphine at m/z 353, and putative N‐methylpachysiphine at m/z 367. The NMT gene candidate, Tel01G.16930, was transiently overexpressed in tobacco leaves in the presence of tabersonine or pachysiphine and reaction products were analyzed by LC–MS and compared to a GFP negative control.
(b) Screening of O‐methyl‐transferase activities with 16‐hydroxytabersonine produced by the TeT16H from tabersonine. OMT candidates are arranged according to neighbor‐joining phylogenetic tree classification (100 bootstrap replicates). Metadata associated with bootstrap nodes are represented by a circle whose fill color scale proportionally with the bootstrap value (lower value, purple; higher value yellow) while branch lengths scales proportionally with substitutions per site value. The scale bar represents 0.1 substitutions per site. Positive differential mass chromatograms of tabersonine at m/z 337, 16‐hydroxylated tabersonine at m/z 353 and the O‐methyltransferase assays with OMTs 16OMT‐like candidates at m/z 367 compared to a 16‐methoxytabersonine standard (pink line). OMT gene candidates and TeT16H were transiently co‐overexpressed in tobacco leaves in the presence of tabersonine and reaction products were analyzed by LC–MS and compared to a GFP negative control. GFP, green fluorescent protein; LC–MS, liquid chromatography coupled to mass spectrometry; m/z, mass to charge ratio; OMT, O‐methyltransferase; pachy., pachysiphine; Tab., tabersonine; TeT16H, tabersonine‐16‐hydroxylase from T. elegans.
